Introduction
The human population, which is presently about 8 billion, has been projected to rise drastically to 10.4 billion by the end of 21st century (www.ourworldindata.org). Although this is a significant milestone for the planet Earth, there is still much work to be done to balance the exponential growth in population and human requirements for clothing, food, water, and safety. In the last fifty years, significant progress has been made in science and technology. Emergence of new field of research in molecular biology and molecular genetics collectively referred to as “Omics” has created endless possibilities for biotechnological applications in various aspects of human life. The impact of biotechnology is already being felt, particularly in the development and production of effective vaccines against communicable and non-communicable diseases. Also, through innovative ideas, coupled with advancements in modern biotechnology techniques, some major food crops have been made healthier by the alteration of the quality or content of their main nutrients (Zhao et al., 2021). In the coming years, the impact of climate change in agriculture will become more aggravated, especially in Africa, where the yield per unit area of crops grown is already the lowest globally (Kyetere et al., 2019). Thus, application of plant biotechnology techniques has been recommended for improved food productivity, through the acceleration of the development of new crop varieties with better capacity for high yield (Lloyd et al., 2023).
This paper discusses the economic impact of biotechnology sector in developed and developing countries around the world, explaining the strategies being used in places where it is being fully explored. Although biotechnology is still perceived by many as an emerging technology that poses a risk to human health and environment, progress made so far in adopting its tools as catalyst for food security in Africa were examined as well as policies put in place by many countries in the world, especially in Africa to regulate crops developed through its application (AUDA-NEPAD APET Genome Editing Policy Framework, AAGEPF, 2022). It is our opinion that certain factors, limiting Africa from reaching its full potential in agricultural production can be addressed by smart combination of biotechnological and conventional breeding approaches. To attain food sufficiency in Nigeria, we highlight key areas where biotechnological techniques can be deployed to increase production of important crops in the country. An instance of a campaign against the development of genetically modified crops in Nigeria and possible solutions to prevent the re-occurrence of such in the future was presented and discussed. In light of the aforementioned, this review uses Nigeria and the possibility of improving its common crops using this technology as a case study to demonstrate how cutting-edge biotechnology, such as genetic engineering and genome editing, can be used to expedite the process of ensuring food security in the continent of Africa.
Biotechnology and food security in Africa
The global market value of biotechnology stands at 295 billion USD. In 2019, the industry successfully employed approximately 900,000 people (Martin et al., 2021). The area has given rise to four well-established technological paths: industrial, medicinal, agricultural, and environmental biotechnology. These sectors are mainly concentrated in developed countries, with USA leading in terms of investment. Since the year 2005, there has been a constant global increase in agricultural biotechnology. Africa, through the African Union (AU), has noticed rapid advancement in biotechnology and has made efforts to secure access to emerging technology in the field. This was demonstrated when the organisation constituted the African Panel on Emerging Technologies (APET) in 2016. Some of the roles of the high level-APET is to provide advice on most the rational approach, strategy and policy regulations for emerging biotechnological techniques in Africa (AAGEPF, 2022) and to advise the union and member states on how to harness emerging biological technology towards agricultural productivity and economic development of the continent.
Africa is considered the epicentre of anaemia and micronutrient-deficient people in the world, with children under the age of five years, adolescent girls, and women being mostly affected. Many people on the continent have become food insecure because of conflicts, cultural discrimination, extreme weather events, as well as poverty and economic shocks (Stevens et al., 2022). This situation has highlighted the failure of current economic models at address the development challenges that the developing world faces. Concomitantly, some other challenges, associated with natural resource constraints, such as insufficiency of water and arable land, which in turn has resulted in an increased rate of unemployment, poverty, and inequality are also being experienced in this part of the world (Oxfam, 2019; Harris-Fry et al., 2020; UNICEF, 2023). Particularly, the poor in rural areas are the most vulnerable and affected, with approximately 88.4 million Nigerians living in extreme poverty and under-development (Statista, 2024).
In 2019, Africa spent 43 billion USD on food importation, which is forecast to hit 90 billion USD by 2030. Decline in per capita food production on the continent is partly due to population explosion which is not matched up with adequate food production. This has resulted in widening the gap between food production and the associated consumption, according to Africa Common Position on Food Systems Food Security (ACPOFS, 2021). Currently, Nigeria is a food deficit nation, spending 10 billion USD annually on food importation to feed its ever-growing population. Although the country leads globally in the production of crops like cassava, yam, and taro, this is mostly due to the annual increase in land area under cultivation but not as a result of improved productivity, in terms of yield/ha (Ikuemonisan et al., 2020; Fufa et al., 2023; Kalu et al., 2023). Even so, post-harvest yield loss (up to 60% in some crops) occurs at various stages of food system, thereby making food unaffordable and unavailable to many (Morris et al., 2019; Ewa, 2021; Businessday NG, 2023). To meet the food needs of its people and ensure food security, Nigeria must embrace novel technology in agriculture and overhaul the food system completely.
Important techniques of biotechnology for food security
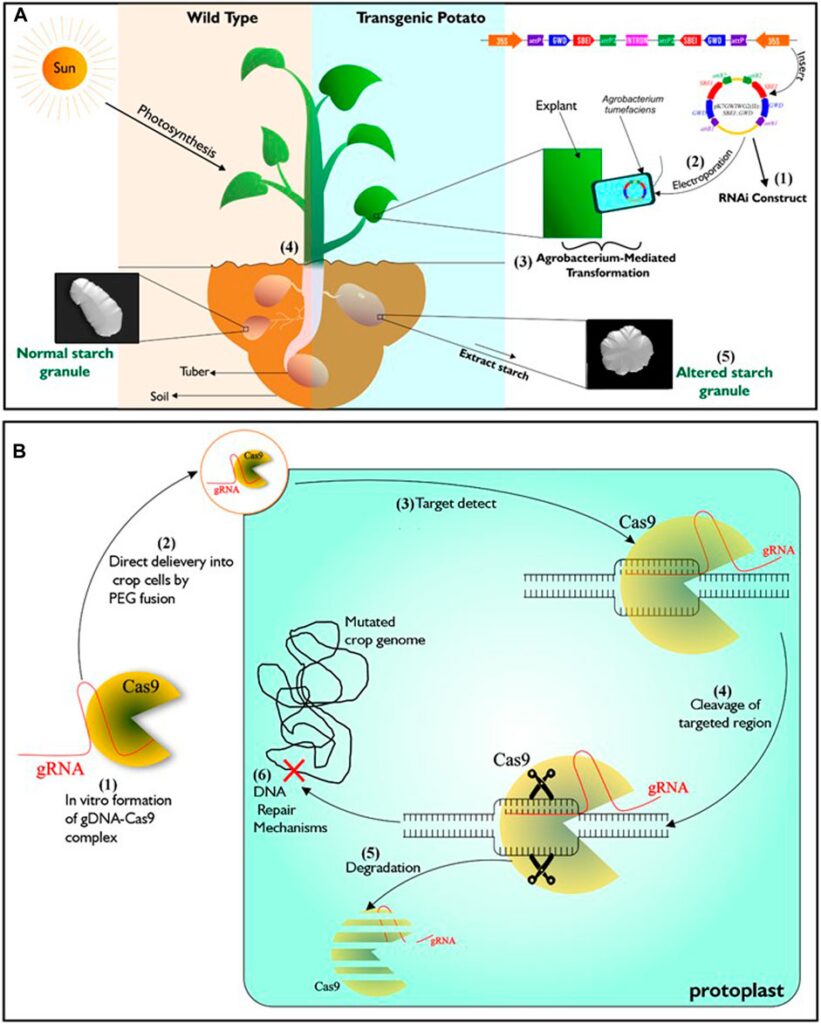
Genetic engineering is one of such advanced technologies that is being utilized to attain food sufficiency in developed countries of the world. It involves exploring knowledge of the functional genomics of species and organisms, by incorporating specific DNA sequences coding for desirable traits into crops of interest (Chen et al., 2017; Narayanan et al., 2019), using tools of gene transfer such as Agrobacterium-mediated transformation, protoplast transformation, electroporation, particle bombardment and calcium-phosphate-mediated gene transformation. This technology can, in addition, bring about silencing (Figure 1A), over-expression or complete loss of function of a specific gene within plants (Brummell et al., 2015; Ferreira et al., 2017; Wang et al., 2018; Zhong et al., 2019). Crops developed through this approach are referred to as transgenic or genetically modified organism (GMO). A meta-analysis of data from the maize field by Pellegrino et al. (2018) over a period of 21 years indicated that genetically engineered maize performed better in grain yield than those from near-isogenic lines.
Figure 1. Illustration of genetic engineering (RNAi) and DNA-free CRISPR/Cas9. (A) Described RNAi gene silencing technology. It starts with the construction of T-Plasmid which contains the coding sequence of the targeted gene/s and antibiotics resistance gene. Agrobacterium tumefaciens is then transformed with the RNAi construct which is then used in transforming young plant tissues such as leaves or internode from tissue cultured wild type. To confirm successful transformation events, putative transgenic lines that are resistant to the incorporated antibiotics resistant genes are selected. Transgenic lines will exhibit phenotypes that is different from wild type depending on the function of the silenced genes. (B) Described DNA-free CRISPR/Cas9 technology. It starts with in vitro complex formation between recombinant Cas9 and gRNA. Then the CRISPR/Cas ribonucleoprotein (RNP) complex is directly delivered to protoplasts by PEG fusion. After delivery into the protoplast, The RNP complex is active and enabled to precisely locate the target genomic region/s to induce double strand breaks. Mutation in the genome occurs after inherent cell repairs at the targeted genomic regions without the addition of foreign DNA. Subsequently, complete degradation of RNP complex occurs within the cell.
Genome editing (GEd) technology is another aspect of biotechnology currently experiencing growth. It enables scientists to manipulate the genome (the entire genetic information found in a cell) of various living organisms. It is based on the use of engineered sequence specific nucleases (SSNs) to induce DNA double-stranded breaks at precise locus/loci in the genome, followed by repair through either of the two cellular DNA repair pathways, including error-prone non-homologous end joining (NHEJ) or homology directed repair (HDR) (Wyman and Kanaar, 2006; Malzahn et al., 2017). Zinc finger nucleases (ZFNs) (Kim et al., 1996), homing endonucleases (HE) or Meganucleases (MN) (Chevalier and Stoddard, 2001) and transcription activator-like effector nucleases (TALEN) (Boch et al., 2009) are the three distinct SSNs, engineered prior to the emergence of CRISPR-CaS system, the most recent technique and by far the most popular tool for genome editing (Wiedenheft et al., 2012). Unlike transgenic plants, a major feature of the genome editing is the ability to alter a plant’s genome without incorporating foreign DNA (Figure 1B; Malzahn et al., 2017). Application of genome editing in crop improvement is favoured for its cost-effectiveness, ease of use and possibility of speeding up the development of elite varieites.
Regulation of GMO and GEd crops and products
There are still unanswered questions about the safety of genetically modified crops for humans and the environment, despite efforts to maximize the benefits of genetic engineering for food security. Because of this, most nations have implemented or are implementing regulatory frameworks that cover the creation, processing, and use of genetically modified organisms (GMOs), though the specifics of these frameworks vary from one place to another. This is because regulators in diverse countries either take a product-based approach or process-based approach for safety regulation of GM crops. A product-base approach assesses risks and gains of GM crop on a case and case basis, a process-based approach the assesses method used in producing GM crops. For example, regulators of GMO in the EU, Australia and New Zealand are concerned about uncertainties in the technology and have refused to approve most GMO products. These items are labelled on the shelf even after they are approved, giving customers flexibility in their selection. This is in sharp contrast to GMO regulation in countries like the United States, Japan, Mexico, and Canada, where regulators ruled that the technology is safe, hence, the approval of products obtained there from for production and consumption. Labelling of GM products in these countries is only done when the nutritional and compositional content is altered or possesses new allergens (Buchholz and Collins, 2010).
The debate over crop enhancement through the use of genome editing (GEd) technology is growing in popularity, as is the use of its products. The public perception did not favour the utilization of GMOs and its regulations are very strict in many countries. Most countries employ science-based risk analysis in regulating such products. The rational approach proposed by scientists for regulating GEd crops is one which designates as GMO, any plant with foreign DNA inserted into its genome whereas any, with no insertion of such is regulated in the same manner as variety developed through conventional breeding methods (Lema, 2019). For example, the approach for regulating GEd crops by biosafety regulators in countries like Australia, Argentina, Brazil, Chile, Canada, United States and Japan, is the same as those that apply to conventional varieties, if foreign DNA or genes are not integrated into the genome (Lema, 2019; Hundleby and Harwood, 2022). Regulations, adopted by Australia for GEd plant is similar to that in Argentina. However, in New Zealand, despite the ruling on environmental protection authority that GEd plants lacking foreign DNA should not be regulated as GMOs in 2014; this stance was later revoked by High Court and currently, GEd crops are categorised as GMOs (Fritsche et al., 2018).
In Africa, Nigeria was the first country to make the move to amend its biosafety legislation to include regulation of genome-edited products (AAGEPF, 2022; AUDA-NEPAD APET Genome Editing Policy Framework, 2022). Thereafter, the guidelines for regulating GEd products were clearly stated and have been adopted since 2020. Other African nations, including Burkina Faso, Ethiopia, Ghana, Kenya, and Malawi, followed suit and enacted national rules for genetically engineered products. The named countries, however exempted regulation of genome-edited crops with no foreign DNA from their biosafety laws. The move is adjudged positive as it will facilitate increase in agricultural productivity of these countries. South Africa still classifies all GEd plants as GM crops, in contrast to other African nations like eSwatini, Senegal, Mozambique, Namibia, Rwanda, Togo, Zambia, Zimbabwe and others who have expressed interest in creating laws for genome-edited products. However, experts opine that such approach contradict the principle of science-based risk analysis and the decision is being appealed (The conversation, 2022).
Involvement of governments in plant biotechnology
The United States of America has continuously led the world in biotechnology research and development as well as the commercialization of its goods for more than 20 years. The United States government implemented regulations in the early years of biotechnology to encourage university-based biotechnology research and development. In 1986, US government enacted the Federal Technology Transfer Act, to foster transfer of publicly developed techniques in biotechnology to private-enterprise (Mugabe et al., 2002). The law made room for joint research between federal laboratories, universities, and private-enterprises and at the inception of innovation, the private partner acquires the patent rights while the participating university and/or government innovators share royalties from licensed innovation. As far back as 1987, the US government spent 2.7 billion USD as funding for biotechnology research and later increased to three billion USD annually by mid 1990s (Avramovic, 1996).
Early investment in R&D and good policy may have encouraged private-sector participation in development and commercialization of biotechnology products and may also have been responsible for huge revenue now generated from their sales. Presently, the US generates a revenue of 33 billion USD from its 318 biotechnology companies (Martin et al., 2021). Because the industry is extremely technical, US government is still committed to funding R&D and training in biotechnology. However, the burden of financing scientific research in the field seems to have shifted to the private-enterprises, as 70% of the funding now comes from that sector. This trend has also been reported in other countries like France and Japan (Martin et al., 2021), an indication that biotechnology sector is highly profitable.
Various developmental strategies were deployed by other countries towards advancement of biotechnology. For example, Brazil gained international prominence in biotechnology after the successful sequencing of the genome of Xylella fastidiosa, a pathogen that causes losses of ∼100 million USD in citrus industry. In 1980, a small team of scientists in Cuba produced alpha-interferon within 42 days. This was a major achievement at the time, which instigated Cuban government in 1986 to fund establishment of centre for Genetic Engineering and Biotechnology. Host of other centers which specialized in biomass conversion, animal production and tropical medicine were also established. As at 2015, biotechnology products, mostly pharmaceuticals, ranked second in the list of most important export commodities in (cuba-solidarity.org.uk 2015; León-de la O et al., 2018). South Korea is another country that has become a biotechnology giant. In 1993, their evolution began, when their government developed a national biotechnology plan, to be executed in three phases, with a total investment of 15 billion USD to be provided by both public and private sectors. Their target was to achieve five percent global market share for novel biotechnology products by 2007. In 2001, the Nigeria Government developed the National Policy for Biotechnology which entails biotechnology knowledge acquisition and commercialization, research and development, capacity building, bioresources development, collaboration in bioresources and biotechnology development. This led to the establishment of National Biotechnology Research and Development Agency (NABDA, 2001) in the same year, which is saddled with the responsibility of implementing the national policy on biotechnology. At the time, strategies for implementing the policy included the identification of Sheda Science and Technology Complex SHESTCO at Federal Capital Territory. In addition, premier universities in the six geo-political zones of the country, namely, University of Ibadan in the southwest; University of Nigeria, Nsukka in the southeast; University of Port-Harcourt in the south-south; Ahmadu Bello University in the northwest; University of Maiduguri in the northeast and the University of Jos, in the northcentral were brought on board to provide necessary facility to support research in biotechnology and genetic engineering. Furthermore, a committee, comprising of representative from Ministries of science and technology, agricultural and rural development, environment health, education, etc., directors of research institutes, universities, manufacturer association of Nigeria and National association of Chambers of Commerce Industry, Mine and Agriculture (NACCIMA), was set up to provide technical expertise needed for biotechnology policy implementation in the country. Currently, the strategy for biotechnology integration in Nigeria has not changed much as indicated in (Figure 2). National Biotechnology Policy was targeted towards accelerated technological growth and increasing self-reliance by strengthening capacity of home-based researchers to copy and adapt techniques in biotechnology for national development. Also, it is expected to serve as government blueprint to effectively address its concerns such as food security among others.
Figure 2. An overview of the biotechnology strategy in Nigeria. Many government agencies and ministries are involved in setting up, implementation of biotechnology policy and regulation of biotech products in Nigeria. Those that are most relevant in the implantation and regulation of biotechnology are National Biotechnology Development Agency (NABDA), National Agency for Food and Drug Administration and Control (NAFDAC), National Biosafety Management Agency (NBMA), Federal Ministries, Nigerian Association of Chambers of Commerce, Industry, Mines, and Agriculture (NACCIMA) and The National Centre for Genetic Resources and Biotechnology (NACGRAB), Sheda Science and Technology Complex (SHESTCO). Research institutes and Universities are involved in human and capacity building, the national Vaccine governing committee consisting of research institutions and federal ministry of health are involved in vaccine research, development, and production in Nigeria.
Since its founding, NABRDA has coordinated biotechnology-related activities, leading to commercialized products and promoted agricultural biotechnology across the country; However, in 2015, Nigeria passed a biosafety bill, which gave the National Biosafety Management Agency (NBMA) control over biotechnology activities in the country. NBMA had the responsibility of ensuring safe application, handling of genetically modified organisms and products through licence issuance in Nigeria. In 2015, SHESTCO, in collaboration with WAAPP/PPAAO and ARCN, organized workshop for young and upcoming bio-scientists, drawn from various research institutions in the country, to strengthen biotechnology capacity in the country. Similarly, the biotechnology research and development centre at Ebonyi State University, Abakaliki has commenced summer training on tropical plant biotechnology since 2013. Cross River Government, a state in Nigeria, by way of public private participation in biotechnology promotion, partnered with a biotechnology company to jump start production of feedstock on a large scale for its juice processing plant in Calabar free Trade zone.
Prospect of using plant biotechnology for improvement of major cropsRice
Rice is the single, most significant cereal crop consumed globally, simply because it is a convenience food for the growing world population, with the demand for it expected to increase by 25% by 2030 (Naik et al., 2022; Poutanen et al., 2022). The crop has also been developed into a model monocot system for genetic and functional genomic research (Datta, 2004; Jung et al., 2008). Although, Africa has over 130 M ha of arable land that is suitable for rice production, only approximately 10 M ha of it is currently used for that purpose (IRRI, 2024), which indicates that Africa’s potential in rice production is still largely untapped. For example, Nigeria, currently the largest producer of rice in Africa, has a potential land area of about 4.9 million hectares suitable for rice production, but has limited cropping to about 1.7 million ha, with production being constrained by low input, poor crop management techniques and water scarcity (Abiwon et al., 2016). As a result, of the seven million metric tonnes of the crop consumed in the country in 2022, over 1.5 million metric tonnes were imported to meet the shortfall in rice production (The conversation, 2023).
Two cultivated species of rice, Oryza sativa (Asian rice) and Oryza glaberrima (African rice) are cultivated in Africa (Van Andel, 2010). Asian rice has been steadily cultivated throughout the world, due to its higher production than African rice, according to productivity assessments. However, because of rich reservoir of genes for resistance to several biotic and abiotic stresses, African rice can thrive better in harsh environment (Wambugu et al., 2013). In certain part of Burkina Faso, Rice farmers prefer cultivation of African rice over Asian rice, due to its resilience against severe environments. Previous efforts to create a rice variety with the high yielding potential of Asian rice and resistance traits of African rice led to the development of New Rice for Africa (NERICA) varieties, which were created by combining traditional breeding techniques with biotechnological approaches. However, African rice is still unrivalled by NERICA in adaptability traits like weed competitiveness (Agnoun et al., 2012). This could be because only around 9% of the O. glaberrima genome is present in the genomic content of NERICA variants (Ndjiondjop et al., 2008). Additionally, O. glaberrima also possesses valuable genes which can contribute to improvement of nutritional quality of rice. For example, the reports of Gayin et al. (2017) and Wambugu et al. (2019) showed that starch granules, which accumulated in the grain endosperm of African rice, has higher amylose content than its Asian counterpart through structural analysis. This indicates that African rice, with its high amylose content, may be a natural source of resistant starch with potential health benefits, particularly in the treatment of type 2 diabetes, a nutrition-related non-communicable disease that has been increasing in Africa recently (Huang et al., 2018).
Contrary to the view of Sarla and Swamy (2005) that O. glaberrima should be used as genetic resource material for improving O. sativa, it is opined that its yield can be improved directly, in addition to other valuable agronomical and nutritional traits. Grain shattering, frequently caused by lodging, is the major cause of low yield of O. glaberrima (Futakuchi et al., 2008). Evaluation of four O. sativa cultivars and 20 accessions of O. glaberima revealed that yield reduction of the later was due to grain shattering (Ndjiondjop et al., 2018) and this indicates that improving African rice for resistance against seed shattering will improve harvesting efficiency, thereby making African rice less inferior to O. sativa. Recently, a novel gene called Seed Shattering 11 (SH11) in African rice was cloned and characterised (Ning et al., 2023). This gene encodes a MYB transcription factor which inhibits the expression of genes involved in lignin biosynthesis. It was shown that a single nucleotide polymorphism mutation in the coding region of SH11 increased the binding ability to the GH2 promoter and consequently reduced the lignin content in O. glaberrima. However, CRISPR-CAS9 mediated knockout of SH11 reduced seed shattering significantly in O. glaberrima (Ning et al., 2023). This indicates that this gene will be a good target for reducing susceptibility of African rice to grain shattering.
Africa faces a problem with food insecurity that goes beyond a shortage of high-yielding cultivars and high demand. The soils in the majority of the farm land used for crop production have been farmed for a long time, making them naturally less fertile. Thus, one of the main obstacles to rice production in Africa is soil nitrogen deficiency, which forces rice producers to rely largely on inorganic fertilizers. The use of inorganic fertilizer indiscriminately contributes significantly to the acceleration of global warming by releasing nitrous oxide into the atmosphere (AATF, 2024). To curb this, scientists in Africa, led by African Agricultural Technology Foundation, have developed Nitrogen Use Efficiency (NUE12) rice variety under the NEWEST Rice project. NUE12 is a transgenic event which involved insertion of barley’s alanine aminotransferase gene (HvAlaAT) into the nuclear genome of NERICA-4. The transgenic variety significantly out-performed Wild type (NERICA 4) in terms of yield at varying levels of Nitrogen application and as such, farmers can opt for lower cost (50%) of N fertilizer while maintaining the yield or the same quantity of nitrogen and increase yield. Three countries, namely; Uganda, Ghana and Nigeria are currently implementing processes which may eventually lead to the approval of NUE12 for commercial cultivation by the farmers in the named countries.
Sorghum
Sorghum [Sorghum bicolor (L.) Moench] is a tropical crop that is native to Africa and grown in several countries of the world. It is well-suited to semi-arid tropics because it is a hardy crop that can withstand harsh and water-scarce environments. According to FAOSTAT, (2020), sorghum production area in Africa is about 27.29 million ha, with total and average grain yield of 27.47 million metric tons and 1.01 ton/ha respectively. Nigeria and Ethiopia are the second and fourth largest producers of sorghum in the world, with United State occupying the top position (FAOSTAT, 2020). The grain is high in starch, which makes it a staple food in developing countries as well as feed for livestock and valuable feedstock for biofuel production in the developed countries. The crop’s comparatively small genome (around 818 MBp) has made it a model plant for researching the genetic components of drought resistance.
In Sub-Saharan Africa, one of the biggest problem of sorghum production is hemi-parasitic weed, belonging to the genus Striga (Muchira et al., 2021), It was estimated that Striga infestation negatively affect the livelihood of 300 million people in West Africa alone (Mboob, 1989). Although farmers often deployed cultural methods in managing Striga, however, genetic improvement of sorghum for resistance to Striga remains the most practical and effective solution. Since Striga is an obligated root parasite, its seeds only germinate when stimulated by chemical signal from the host plant (Rich and Ejeta, 2007). Therefore, low striga germination stimulant activity caused by genetic factors will be a good strategy for controlling yield losses due to Striga infestation (Pérez-Vich et al., 2013). Strigolactones—a group of compounds synthesised by most angiosperms as hormones to regulate branching of shoot and root (Gomez-Roldan et al., 2008; Rasmussen et al., 2012)—is the most potent germination stimulant among the sorghum root exudates (Gobena et al., 2017). A mutant allele at LGS1 (LOW GERMINATION STIMULANT 1) locus that drastically reduce Striga germination stimulant activity was identified by Gobena et al. (2017). This LGS1 is now a targeted gene for editing as Steven Runo from Kenyatta University, Kenya already used CRISPR-Cas9 system to develop a striga resistant variety of sorghum by knocking out SgLGS1. Similarly, mutagenesis of genes which encode Carotenoid Cleavage Dioxygenase—an enzyme involved in the biosynthesis of strigolactones in many plants species (Gao et al., 2018; Dutta et al., 2019)—was knock-out in sorghum by CRISPR system, resulting in significant reduction in Striga germination even though it negatively affected the yield (Hao et al., 2023).
Millets
Millets are a diversified collection of small-seeded dryland cereals that are resilient to harsh climatic conditions, tolerant of poor soil conditions, and do not require excessive fertilizer or pesticide application. There are several types of millets, namely; foxtail millet (Setaria italica), pearl millets or bulrush millet [Pennisetum glaucum (L.) R. Br.], finger millet (Eleusine coracana), proso millet (Panicum miliaceum), kodo millet (Paspalum scrobiculatum), barnyard millet (Echinochloa spp.), browntop millet (Panicum ramosum) and little millet (Panicum sumatrense) (Upadhyaya et al., 2006). Climate change causes terminal moisture stress and erratic rainfall patterns at different stages of crop production across the world, thereby, reducing the yield of popular cereals like wheat and rice. However, millet due to its resilience to both biotic and abiotic stresses, has the potential to maintain a more stable yield with high quality. Hence, together with sorghum, these crops are critical for food security in the Sahel regions of West Africa, where they provide over 75% of the total caloric intake for highly food-insecure people in the region (Ndjeunga and Nelson, 1999).
Millets grains are frequently ground into flour and consumed in the form of porridge. They are also used for fermented drinks and other food purposes. When compared with other major cereal crops such as wheat and rice, millets are more nutritionally superior (Ragaee et al., 2006). The grains are high in various essential minerals-calcium, potassium, phosphorus magnesium, iron, and zinc-and vitamins, that help to reduce malnutrition (Hariprasanna et al., 2014; Elangovan et al., 2022). Also referred to as nutria-cereals or nutria-millets, millets are rich sources of essential fatty acids, proteins, carbohydrates, phytochemicals, and antioxidants. Millet grains are rich in polyphenols and particularly, finger millet starch has more amylose to amylopectin ratio, which confers several health benefits on the crop such as; reducing the risk of high blood pressure and heart diseases, and can help in the management of type 2 diabetes and obesity (Swaminaidu et al., 2015; Kumar et al., 2018).
Despite the innate ability of millets to cope with both biotic and abiotic stresses, the scourge of climate change is expected to be more biting on agricultural productivity in the future, leading to low yield and poor grain quality. Therefore, there is a need to further boost the resistance of millets against diseases caused by pathogens, combined stress caused by severe drought and excessive heat, and modification of plant architecture to prevent lodging and bio-fortification with essential micronutrients.
In Africa, pearl millet is the most cultivated, constituting 75% of annual millet production with the rest made up mostly by finger millet. However, unlike other cereals, pearl millet grains are particularly high in lipid content (Sharma et al., 2015). After milling of its whole grain, endogenous lipases are released and cause rapid onset of hydrolytic rancidity. This phenomenon makes the flours of most pearl millet varieties unstable 5–7 days after milling (Aher et al., 2022). Thus short shelf-life of pearl millet flour was identified as the major reason why pearl millet has remained unpopular (Goswami et al., 2020).
Generally, millets are regarded as orphan crops because previously, it has received less attention in terms of genetic studies and crop improvement from the scientific community. This is exacerbated by a lack of efficient transformation systems to successfully develop transgenic varieties or induce mutation in the genome through genome editing techniques because millets are recalcitrant to regeneration via tissue culture methods. In the last decade, millet has however gained attention because of its resilience to harsh climatic conditions and the supreme nutritive value of its grains. For example, the first draft genome of the pearl and finger millet was published by Hittalmani et al. (2017) and Varshney et al. (2017) (Ramu et al., 2023). The annotated genome of finger millets by Devos et al. (2023), is now available on Phytozome https://phytozome-next.jgi.doe.gov/info/Ecoracana_v1_1. It appears the efficiency of millet transformation is determined by factors like the choice of explants and the use of synthetic secondary metabolites like acetosyringone (Sood et al., 2019; Bhatt et al., 2021; Ceasar, 2022). It may also be influenced by specific millet genera being studied. For example, finger millet is predominantly transformed by Agrobacterium, with studies reporting the development of transgenic finger millet (Ceasar et al., 2011; Ignacimuthu and Ceasar, 2012; Satish et al., 2017). On the contrary, pearl millet seems to be transformed mainly by the biolistic method (Sood et al., 2019).
The first Genome editing in millet was reported by Lin et al. (2018) where the protoplast technology was used in creating CRISPR/Cas9 mutagenesis. Also demonstrated was the haploid induction in foxtail millet, by knocking out SiMTL gene using the CRISPR-Cas9 system (Cheng et al., 2021). More recently, Liang et al. (2022) also utilized Agrobacterium-mediated transformation to induce multi-genic mutation in the foxtail genome. A single-base editing in foxtail millet was also first reported in this study and it resulted in the creation of an herbicide-tolerant plant. Presently, it appears that not much success has been achieved in genome editing of millets, there is no report of genome editing of other millets apart from foxtail millet. Since pearl millet is widely cultivated in Africa, efforts must be intensified to address the problem of the short shelf-life of its flour. Recently, Aher et al. (2022) reported that the loss of functional triacylglycerol lipases in the grains of pearl millet is linked with low flour rancidity. This was demonstrated when a mutation in two genes PgTAGLip1 and PgTAGLip2 resulted in the loss of function and was consistent with low flour rancidity in some pearl millet varieties. Therefore, the two candidate genes will be good targets for genome editing to improve the shelf-life of pearl millet flour. Other genes which may be the target of genome editing for improvement of the resistance of millets to abiotic stresses such as drought have also been reviewed by Krishna et al. (2022).
Maize
Maize (Zea mays L.), is considered the most important cereal crop in Central and Eastern Africa. For example, maize contributions to Kenyan economy include the generation of employment, serving as means of livelihood for many families as well as source of food security and foreign exchange earnings. Also, maize is the most important food crop in Kenya as ∼96% of the population consumed the crop as staple food every day (Njuguna et al., 2017). This is an indication that any significant yield loss due to abiotic or biotic stresses on the crop will have a damaging effect on food security in Kenya. An example of biotic stress that has caused significant yield loss in the country in recent time is Maize Lethal Necrosis (MLN), a viral disease that has caused between 23%–100% yield loss, estimated to be 180 million USD (Redinbaugh and Stewart, 2018). Conventional method for breeding high yielding maize varieties that are resistant to MLN is preferred over the use of pollution-prone method involving spraying of the MLN vector (Aphids and Thrips) (Awata et al., 2021). However, introgression of MLN resistant genes into susceptible varieties through backcrossing will take years to develop and may still suffer yield penalty as a result. To bypass this challenge, Kenya Agriculture and Livestock Research Organization in association with four other international partner organisations, embarked on genome editing project which seeks to identify and introduce MLN resistance genes into elite varieties (e.g., CML536) that are susceptible to the disease (CGIAR, 2022; Table 1). Since 2021, the MLN susceptible gene in elite varieties has been identified on maize chromosome 6 and has been edited to its resistance form against MLN disease. The role of the edited gene which now confers resistance on otherwise susceptible lines has been validated.
Table 1. Africa’s on-going and successful applications of genome editing for crop improvement.
Drought is another factor that restricts maize output in Kenya in addition to other abiotic stress. This is because about 75% of the available land area in the country is arid and semi-arid (Zeila and Jama, 2005). Exposure of plants to drought stimulates overproduction of reactive oxygen species and increased oxidative stress which causes damage to primary metabolites (DNA, protein, Lipid proteins, and carbohydrates), cell death and loss of whole plant (Njuguna et al., 2017). The impact of drought on maize is phase specific, for example, when it occurs before anthesis, maize undergoes delay in flowering (Abrecht and Carberry, 1993), whereas if it is at grain filling stage, it can cause a more devastating effect like low grain yield (Schussler and Westgate, 1995). Recently, Njuguna et al. (2017) used CRISPR-Cas9 to knock down poly (ADP-ribose) polymerase (PARP), a gene that plays a major role in the maintenance of the energy homeostasis during stresses and significant increase in tolerance to oxidative stress resulted.
In 2016, there was an outbreak of fall armyworm (FAW; Spodoptera frugiperda) in Nigeria and São Tome, leaving large scale destruction in its trail (Goergen et al., 2016). This invasive insect has since spread to eastern African countries like Ethiopia, Kenya and Tanzania (Sisay et al., 2019). Spatial assessment of climate suitability of FAW indicated that Africa is generally favourable for this insect with the exception of Lesotho and South Africa (Senay et al., 2022). Moreover, most of the maize varieties are also susceptible to seasonal infestation of FAW. To control crop pests, three possible solutions exist; use of insecticides, integrated pest management (IPM) and genetic improvement of maize for resistance against FAW. Genetic solution appears to be the best option because maize production is largely done by smallholder farmers who cannot afford to spray their farm numerous times and IPM is not widely practiced in Africa (Alwang et al., 2019).
The TELA Maize Project led by Kenyan-based African Agricultural Technology Foundation, coordinated the insertion of a Bacillus thuringiensis gene which encodes Cry2Ab delta-endotoxin (Table 2), into the genome of drought tolerant maize varieties, thereby conferring insect protection and drought tolerance in TELA® maize varieties through conventional breeding and biotechnology methods. Cry2Ab delta-endotoxin is a toxic protein which has lethal effects on the digestive system of lepidopteran and some dipteran insects (McNeil and Dean, 2011), however, it is not harmful to human and livestock, having been used in organic farming for over half a century to control insect pests. In a confined field trial of TELA® maize in Nigeria by Oyekunle et al. (2023), it was reported that the transgenic TELA® maize genotypes were resistant, not only to FAW but even to stem borer—another destructive pest of maize. Five TELA® maize hybrids have been cultivated on a commercial scale by South African farmers since 2016 (Table 2). Also, Nigeria government, through the National Varieties Release Committee, in January 2024 approved the release of the seeds to of four TELA® maize varieties (SAMMAZ 72T, SAMMAZ 73T, SAMMAZ 74T, and SAMMAZ 75T) to farmers and commercial production in the country (Alliance for Science, 2024). In Kenya, farmers awaits the release of three TELA® maize hybrids (WE1259B, WE3205B and WE5206B) as it has been recommended by the Kenya Plant Health Inspectorate Service through National Performance Trial Committee (KALRO, 2024).
Table 2. Highlights of genetic engineering projects in various African countries.
Cassava
Cassava is a major crop in the tropical and subtropical regions of the world, cultivated for its starch-rich swollen roots (Okogbenin et al., 2002; Okogbenin et al., 2003; Okogbenin et al., 2012). Agronomical qualities like resilience against stress-prone environments, adaptability to subsistence farming systems and high starch content of its storage roots, makes it the crop of choice for millions of smallholder farmers (Montagnac et al., 2009; Howler et al., 2013). Harvested storage roots are processed and converted to various food products in sub-Sahara Africa (SSA), providing over 50% of the caloric intake of one-third of the entire population (Okogbenin et al., 2007; Ewa, 2021). Nigeria is the world’s leading producer of cassava (Table 3). However, the country is still not at its full potential in terms of yield (tonnes/hectare), as it produces FAOSTAT, 2017). In fact, over 20% decline in yield/hectare of storage root was reported between 2007 and 2017, despite a significant increase in the area of land cultivated within this period (Otekunrin and Sawicka, 2019). Factors, such as susceptibility to diseases and high post-harvest loss due to rapid physiological deterioration are major limitations preventing Nigeria from reaching its full potential in cassava production (Fregene et al., 1997; Fregene et al., 2000; Akano et al., 2002; Ogbe et al., 2006; Akinbo et al., 2007; Akinbo et al., 2011; Akinbo et al., 2012a; McCallum et al., 2017; Zainuddin et al., 2018). However, with biotechnology approach, scientists from national agricultural research institutions in Brazil, Nigeria, Ghana and Uganda, are rapidly building resistance to green mite, whitefly, cassava mosaic disease and post-harvest physiological deterioration (PPD) with the support of their counterparts at the International Center for Tropical Agriculture (CIAT) in Colombia (Fregene et al., 1997; Akano et al., 2002; Rudi et al., 2010; Akinbo et al., 2012b).
Table 3. The total land area, production of major crops in Nigeria and export quantity.
Other factors that contribute to final yield of cassava which are currently being worked on for improvement include storage root yield and starch content (Okogbenin et al., 2003). Significant progress has been made in this area since Cassava Source-Sink (CASS) project began. Funded by Bill and Melinda gate foundation, CASS project focuses on combining techniques in plant biotechnology with physiological processes to develop cassava genotypes with increased storage root and starch yield. The goal of the project is to boost the income of smallholder farmers in SSA (Sonnewald et al., 2020).
Due to heterozygosity of the crop, simultaneous stacking of multiple (desirable) genes in a cassava variety through plant breeding methods is very challenging (Fregene et al., 2006; Beyene et al., 2018). However, biotechnological approach has been utilized to overcome this (Fregene et al., 1997). The technique is considered as the most reliable option for combining multiple traits in a crop. For instance, CASS project adopted systematic gene stacking approach, involving simultaneous incorporation of foreign genes for increased storage root and starch yield (Sonnewald et al., 2020). To this effect, several transgenic lines are currently under confined field trial (CFT) in National Chung Hsing University, Taiwan and International Institute of Tropical Agriculture Ibadan Nigeria (Sonnewald et al., 2020; CASS, 2023).
Although, cassava is regarded as a food security crop, meals made from its swollen storage roots are considered unwholesome, due to the absence of essential minerals such as iron and zinc in the crop (Sayre et al., 2011). Deficiency of zinc and iron is common among children under the age of five and pregnant women in Nigeria (WHO, 2008; NMHN 2005; Table 3). Genetic engineering method has also been utilized in the biofortification of the crop by co-expression of iron transporter and ferritin genes from A. thaliana which results in accumulation of iron and zinc in the storage root, up to a substantial level in human diet (Narayanan et al., 2019). Similarly, a transgenic cassava variety developed by Beyene et al. (2018) not only shows high pro-vitamin A in its storage roots but also exhibited longer shelf life when compared with ordinary variety. Taylor et al., 2004; 2012; Siritunga and Sayre 2003; Zhang et al., 2005 similarly worked on other traits to improve cassava qualities.
Since genome editing of Cassava was first reported by Odipio et al. (2017) using CRISPR/Cas9 system, the technique have been effectively utilized to control high accumulation of toxic compounds in the leaves or roots of cassava. For example, Juma et al. (2022), successfully lowered cyanide levels in cassava by inducing mutagenesis in CYP79D1 gene—encodes Valine N-monooxygenase 2 which catalyses major reaction in the synthesis of cyanogen in cassava—using CRISPR/Cas9 system. Similarly, other traits such as herbicide tolerance, modification of root starch structure and disease resistance have been possible through CRISPR/Cas9-mediated targeted mutagenesis (Bull et al., 2018; Hummel et al., 2018; Gomez et al., 2019; Veley et al., 2021).
Yam
White yam (Dioscorea rotundata) is the most popular species among tuber-bearing crops in the family Dioscoreaceae. According to FAOSTAT 2021, Nigeria was the largest producer of this crop in the world. Its economic value is derived mainly from the sale of its starch-rich tuber and day-to-day consumption, providing about 285 dietary calories for 300 million people in Sub-Saharan Africa (Adejumo et al., 2013). Apart from its rich starch deposit, yam tuber also contains higher a vitamin C and crude protein content than the swollen storage roots of cassava, making it a healthier food source (Baah et al., 2009). It is also an important source of secondary metabolites such as steroidal saponins (diosgenin), diterpenoids, and alkaloids, which are utilized in the manufacturing of pharmaceutical products (Mignouna et al., 2008; de Lourdes Contreras-Pacheco et al., 2013).
A major constraint to yam production is infestation with parasitic nematodes, insects, fungi, and viruses, which reduce both the yield and quality of tubers (Asante et al., 2008; Asala et al., 2012; Zaknayiba and Tanko, 2013). Amusa et al. (2003) claimed that nematode infestations might also result in a sizeable yield loss. Despite Aighewi et al. (2015) reporting that viruses can cause a 50% yield loss. These problems have become more severe lately due to the repetitive usage of pest-infested tubers, usually from previous cropping seasons for planting with each successive year (Amusa et al., 2003; Aidoo et al., 2011). Inter-state exchange of these infected planting material further worsens the situation, as mixed viral infections were detected in a survey carried out in Guninea-Savana. Zone of Nigeria where yam is mostly produced (Asala et al., 2012). As such, Aighewi et al. (2015) recommended the use of planting material developed by tissue and organ culture as a better alternative to the pervasive traditional way of yam propagation, stating that it is fast and guarantees the production of disease-free planting material. Also, yam clones can be cleaned from virus disease by cryotherapy, a method which involves short-term treatment of the infected samples in liquid nitrogen to eradicate the pathogens (Wang and Valkonen, 2009). Eradication of bacteria, viroids, and viruses using this method have been reported in the planting material of economically important crops such as banana (Helliot et al., 2002), potato (Wang et al., 2006) and sweet potato (Wang and Valkonen, 2008).
Transfer of useful genes from wild families of yam to elite varieties has proven difficult (Spillane and Gepts, 2001) because yam improvement through conventional breeding methods is constrained by poor flowering, polyploidy, and heterozygosity (Mignouna et al., 2008). However, a protocol for Agrobacterium-mediated transformation of yam has been developed (Nyaboga et al., 2014), and this has opened a new opportunity for developing yam varieties that are resistant to viral and nematode infection through genetic engineering, although optimization of this protocol for different yam genotypes may be necessary for effective and increased transformation efficiency. Additionally, knowledge of nematode-host interactions could be explored through a biotechnological approach to target yam-nematode activity by disrupting the nematode’s infective stage which may reduce passage potency through yam tissues, fecundity, establishment in yam cells and/or feeding ability on susceptible yam cultivars. In a study by Jaouannet et al. (2013), repression of calreticulin gene expression in Meloidogyne incognita by RNAi–RNA interference led to a decrease in the ability of the nematode to infect Arabidopsis thaliana and overexpression of M. incognita calreticulin gene in A. thaliana increased the susceptibility of the plant to the nematode infection. The study clearly shows the role that biotechnology can play in manipulating host-pathogen interactions. Fosu-Nyarko and Jones, (2015) suggested that RNAi technology possesses great potential in conferring resistance to several nematode species on the plant, unlike the introgression of natural resistant genes. Henceforth, genome editing may also play a key role in improving various traits in yam. The first successful CRISR/Cas9-based genome editing and validation in yam was reported by Syombua et al. (2021). Agrobacterium harbouring the CRISR/Cas9 vector was used in targeted mutagenesis of the phytoene desaturase gene in D. rotundata.
Cocoyam
Cocoyam is often used interchangeably in literature for species of the two most cultivated genera, Colocasia and Xanthosoma, belonging to the family Araceae. The genus Colocasia spp. has between 11 and 16 species (Long and Liu, 2001; CABI, 2013), whereas Xanthosoma spp. has about 60 species (Bown, 2000; Stevens, 2012). Taro, or old cocoyam (C. esculenta) and tannia or new cocoyam (Xanthosoma sagittifolium) are by far the most cultivated species in the world (Ubalua et al., 2016; Degefa and Anbessa, 2017). In West Africa, X. sagittifolium is the main edible aroid and has overtaken Colocasia esculenta, probably because its plant is more robust and tolerant to drought and the cormel, a rich source of starch for domestic and industrial use, is adapted to preparing indigenous foods like fufu (Onokpise et al., 1999; Jennings, 2009). The small granule size of its starch (taro) is easily digestible, thereby making it a healthy energy food for children (Adekiya and Agbede, 2016). Nigeria is the largest producer of cocoyam in the world (FAOSTAT, 2021) and approximately 86.27 × 106 million hectares of arable land are still available to further scale-up the production of the crop in the country (Chukwu, 2015). Despite the enormous potential of cocoyam production in the country, production is limited by a lot of factors, among which, frequent incidence of pests and diseases is a major challenge. In 2009, cocoyam production was nearly disrupted completely because of the severity and pervasiveness of taro leaf blight which attacked farms in Nigeria (Ubalua et al., 2016). Although chemical control of pest is considered an option in the control of diseases, an increase in the prices of pesticides, destruction of the underground cocoyam corm-setts by broad-spectrum herbicides in the farm and the toxicity of some active ingredients in the pesticides have discouraged many farmers from subscribing to this option (Acheampong et al., 2015).
To control pest infestations, developing genetically resistant cultivars is recommended as the safest and most economical option. Improvement by conventional breeding methods is difficult because cocoyam rarely flowers naturally but this can be induced artificially (Tambong and Meboka, 1994; Jennings, 2009). Techniques for pollen storage and germination have been developed to facilitate sexual crossing in the crop (Agueguia and Fatokun, 1987). However, some of the methods for artificial flower inducement may be cultivar-dependent as observed in the cocoyam cultivar “Bun Long” which does not respond to inducement by gibberellic acid (Ivancic et al., 2004). Tissue culture has been deployed in the multiplication of cocoyam cultivars that are resistant to taro leaf blight through collaborative research by the NRCRI and the European Union since 2010. This effort is laudable, as it encourages the development and distribution of resistant cultivar/s among farmers. Traditional cocoyam breeding methods can take up to 10 years, whereas genetic engineering can be used as a substitute technique to produce varieties with highly desirable traits. The Glucuronidase (Gus) gene (Fukino et al., 2000) and rice chitinase gene chi11 (He et al., 2010) have been transformed into cocoyam by particle bombardment, although, transformation efficiency was low. Agrobacterium-mediated transformation of cocoyam (Bun long Cv.) with rice chitinase gene chi11 and wheat oxalate oxidase gene gf2.8 led to the production of a transgenic potato that is resistant to Sclerotium rolfsii and Phytophthora colocasiae (He et al., 2008, 2013). This method appeared to be more promising, because its transformation efficiency is high (between 1% and 3%), opening up a greater opportunity for other crops than what was obtained for the particle bombardment method (Agrobacterium-mediated transformation of Taro has been developed by He et al. (2015).
Groundnut
Groundnut, also known as peanut, is a leguminous crop cultivated in the semi-arid and subtropical regions of the world for its oil-rich seed and consumed as food and feed. Groundnut seed which contains 44%–56% oil is very high in unsaturated fatty acid 85% (Sabate, 2003), making it a top choice among oilseed crops. The protein content of the seed is about 22%–30%, also it is a rich source of essential minerals and vitamins. Groundnuts, with their high seed oil and protein contents, play a crucial role in preventing malnutrition and guaranteeing food security. Frequent nut consumption is associated with lower rates of coronary artery disease. Also, nut-rich diets improve the serum lipid profile of participants in dietary intervention trials. Despite their high-calorie density, groundnuts produce satiety, limited energy absorption, and enhanced energy expenditure after eating, hence they do not significantly contribute to weight gain (Sabaté, 2003; Mattes et al., 2008).
Groundnut haulms, a by-product is not only gaining prominence as a fodder source for feeding livestock during the dry season when green grasses are unavailable (Ajeigbe et al., 2014) but also due to dwindling arable land and water resources occasioned by climate change in the Sahel region of West Africa (Blümmel et al., 2012). Nigeria ranks fourth in the world and first in Africa in groundnut production, contributing about 2.42 million MT out of the 45.3 million MT produced in 2017 (FAOSTAT 2017). Regardless of Nigerian’s ranking globally in groundnut production, the country seems not to be making progress in the production of this crop, for example in 2017, the USA surpassed Nigeria in groundnut production for the first time in over 10 years with a production of 3.28 million MT (FAOSTAT 2017). This may not be unconnected with Africa’s poor groundnut yield/ha, reported to be (929 kg/ha) unlike those obtainable in Asia and America (2,217 kg/ha) and (3,632 kg/ha) respectively (FAOSTAT, 2014).
Additionally, Ajeigbe et al. (2014), identified biotic and abiotic stresses as major factors constraining groundnut production in Nigeria. For example, groundnut is highly susceptible to aflatoxin contamination, which are secondary metabolites synthesised by aflatoxigenic fungi like Aspergillus flavus and A. parasiticus after infecting the pods or seeds at the preharvest and post-harvest stages. According to Wilson and Stansell (1983) and Cole et al. (1995), aflatoxin contamination is an extremely variable trait that arises largely under heat and drought stress, and the location where groundnut is primarily grown in Nigeria is especially prone to these abiotic stresses. Aflatoxins are highly toxic to humans and have been linked to liver cancer, suppression of the immune system, and retarded growth in children (Bhatnagar-Mathur et al., 2015). Several countries throughout the world have been obliged to implement rigorous guidelines for permitted levels of aflatoxins in groundnut imports. As a result, Africa loses around USD 500 million per year in export trade due to systematic rejections of export crops and animal products with unacceptable levels of aflatoxins (Janila and Nigam, 2013).
Competitive atoxigenic fungal technology and deployment of promiscuous atoxigenic Aspergillus are some of the preharvest strategies that have been effectively used to reduce the level of aflatoxin contamination but it poses the problem of compromising the quality of the kernels and hygiene. Thus, the development of varieties that are resistant to the preharvest infestation of groundnut by A. flavus remains a viable option, though it has remained a challenge for peanut breeding programs (Janila and Nigam, 2013; Bhatnagar-Mathur et al., 2015). To effectively minimize the occurrence of pre-harvest aflatoxin contamination, the mechanism initially proposed by Holbrook et al., 2009 and corroborated by Janila et al. (2013) involves the identification of groundnut genotypes that are resistant to either drought or root-not nematode (Holbrook et al., 2009). This was because significant positive correlations were observed between resistance to these stresses and aflatoxin contamination. Subsequently, the potential for enhancing antifungal activities in groundnut seeds using marker-assisted selection was demonstrated by Yu et al. (2020), where SNP marker system was used to identify gene markers in two novel groundnut genotypes linked to genes that confer resistance against aflatoxin contamination.
Various strategies have recently been deployed for the transformation and development of transgenic groundnuts with alteration in the complex interaction between pathogen and groundnut-host system. This involves the use of genes that encode proteins/enzymes (antimicrobial peptides like defensins) which activate defence mechanisms against fungi and aflatoxin or host-induced silencing of Aspergillus genes encoding key enzymes involved in fungal sporulation or aflatoxin production. Overexpression of Medicago sativa Defensin 1 and Medicago truncatula Defensing 4. 2 and through HIGS of the aspergillus gene; AFlM (Ver-1)–encodes versicolorin dehydrogenase–and AflP (omtA)–encodes methyltransferase–resulted in high level of resistance in groundnut to aflatoxin production (Sharma et al., 2018). Similarly, multiplexed HIGS of A. flavus genes (AflM, AflR, veA and nsdC) also enhance resistance to infection caused by Aspergillus and aflatoxin contamination (Prasad et al., 2023).
With the successes recorded so far through various strategies mentioned, the future of genome-edited groundnut with high resistance to aflatoxin is very promising. Firstly, there is abundant information on the genomic information (whole genome sequences and annotations) of A. flavus to study its biology (Payne et al., 2006; Payne et al., 2008; Nierman et al., 2015; Ohkura et al., 2018; Fountain et al., 2020; Bharose et al., 2024). Also, the reference genome of groundnut is available (Zhuang et al., 2019). The first CRISPR-based editing of Fad2-the gene encodes the enzyme that catalysis the conversion of oleic acid to linoleic acid (Schwartzbeck et al., 2001)- in groundnut was reported by Yuan et al. (2019) and it resulted in elevated levels of oleic acid and reduction in linoleic acid for improved oil quality and better health benefits. Neelakandan et al. (2022) went further to create the first induced base editing of FAD2 genes in groundnut, using CRISPR/Cas9.
Sesame
Sesame (Sesamum indicum L.) is among the ancient oil-yielding crops. Its seeds, when decorticated, bear one of the highest oil contents. The oil is made up of 83%–90% unsaturated fatty acids (Fukuda et al., 1985; Anilakumar et al., 2010). The seed is also rich source of protein, vitamins, minerals and lignans (methylenedioxyphenyl compounds like sesamolin, sesamin, sesamol and tocopherols (Fukuda et al., 1985). In 2021, Nigeria was the second largest producer of sesame in Africa and the sixth in the world. In terms of export quantity, the crop was only second to cocoa, suggesting that sesame has high potential for contributing to the country’s foreign earnings (FAOSTAT, 2021). According to Tukura and Ashindo (2019) Nigeria earned 139 million and 1.4 billion USD from exporting sesame in 2010 and 2012 respectively. However, a review of sesame seed production in Nigeria from 2003 to 2012 by Umar et al. (2014) revealed that increase in production experienced within this period was due to increase in land area used for cultivation rather than an increase in average yield per hectare, which was very low and mostly similar to the world’s average of 0.49 t/ha (FAO, 2012). Early senescence and extreme susceptibility to biotic stresses like bacterial blight (Xanthomonas campestris pv. sesame) and powdery mildew (Oidium erysiphoides), and abiotic stresses like photosensitivity and waterlogging are major constraints to increasing sesame yield (Dossa et al., 2017). Even though studies have shown that the wild species of the crop is a repository of desirable genes that can help elite varieties cope with these stresses (Kolte, 1985; Brar and Ahuja, 2008), post-fertilization barriers remain a major hindrance to transferring desirable genes into elite varieties by conventional breeding approach (Tiwari et al., 2011). Thus, genetic engineering is the only available option for the transfer of those useful genes from the wild species into the elite varieties. Since Yadav et al. (2010) reported the first successful Agrobacterium-mediated transformations of sesame, other authors, like Al-Shafeay et al. (2011) and Chowdhury et al. (2014) have also achieved similar results in about 42.66% transformation efficiency. Agrobacterium-mediated transformation of sesame brought about the development of multiple-stress tolerance in the crop by overexpression of Osmotin-like proteins (SindOLP) gene (Chowdhury et al., 2017). Morphological features of the plant, like the number of capsules per plant, the number of grains per capsule, grain weight, plant height, length of capsules, number of capsules per axil and axis height of the first capsule has been associated with grain yield of sesame (Dossa et al., 2017; Teklu et al., 2022). Wei et al. (2015, 2016) stated that problems associated with low yield in sesame production may be solved through functional genomic study involving multigenic assemblage of experimentally determined genes such as SiGA20ox1 and two candidate genes for plant height (SiDFL1) and (SiILR1) in a transgenic sesame line delivered by Agrobacterium-mediate transformation.
Sweet potato
Ipomoea batatas, the common sweet potato, is an important staple food of many tropical and temperate countries, as it ranks fifth in developing nations in terms of economic value and seventh for energy consumption (Loebenstein, 2009). It plays an important role in nutritional improvement, as well as serving as raw materials in the processing of feeds, starches and bioethanol in various industries (Antonio et al., 2011). Africa is the second largest producing region, with almost 17% of the world’s production and more than 42% of the world’s area, mainly for human consumption (Zhang et al., 2018). Globally, traditional breeding has significantly contributed to trait improvement in the crop (Thottappilly and Loebenstein, 2009). Crop variety is the main variable often manipulated by farmers to raise yields. Egeonu and Akoroda, (2010) characterized and evaluated for sequential selection, 125 clones of sweet potato for different end-uses. In recent times, the biofortification of sweet potato with provitamin A carotenoids have proven to be an economical and potentially sustainable strategy to alleviate vitamin A deficiency (VAD) in developing countries (Tumuhimbise et al., 2013). The flesh of sweet potatoes can be white, yellow, purple, or orange in colour. Based on this diversity, it has been linked to acceptance in terms of nutrition and taste. Particularly, orange-fleshed sweet potato (OFSP) types are the most affordable and year-round source of vitamin A available for low-income families (Nkhata et al., 2020). Initially, most orange-fleshed sweet potatoes had lower dry matter content and poor environmental adaptability than ordinary white sweet potato varieties (Mwanga and Ssemakula, 2011). However, several years of breeding has produced OFSP with improved yield, flavor, drought resistance, dry matter content and early maturation, resulting in increased adoption by farmers (Hummel et al., 2018; Jenkins et al., 2018). National Root Crops Research Institute (NRCRI) has come up with the development of 2-orange and 1- white-fleshed sweet potato to improve the nutritional wellbeing of Nigerians. Additionally, purple-fleshed sweet potato clones, capable of accumulating anthocyanin (powerful antioxidant) in their storage roots have been developed (Montilla et al., 2011). Researchers have reported the encouraging health benefits of OFSP intervention into the staple food, currently available in more than three African countries, including Nigeria. Neela and Fanta, (2019) reviewed the detailed nutritional composition (proximate, mineral, carotenoids, vitamins, phenolic and antioxidant property’s role in Vitamin A deficiency (VAD) management and different food products that can be made from OFSP. Scientists at IITA in Nigeria and in Ethiopia have also developed methods to produce virus-free sweet potato plant through meristem culture (IITA, 1989; Dugassa and Feyissa, 2011). Nonetheless, a lot can still be done on sweet potato production. Biotechnological tools, such as gene transfer would be very effective in its improvement, as they will enable direct introduction of desirable genes from pre-adapted cultivars. In addition, selection using DNA markers would accelerate conventional breeding programmes in Nigeria. The whole genome sequence of sweetpotato is yet to be available publicly and this has limited research effort directed at improving its agronomic traits. However, the genome sequence of its two diploid relatives I. triloba and I. trifida has been done and it can still be utilized as reference genome for studying hexaploid sweetpotato (Wu et al., 2018). Targeted mutagenesis of genes encoding key enzymes in starch biosynthetic pathway (GBSSI and SBEII) was the first report of genome editing in sweetpotato (Wang et al., 2019).
Tomato
Globally, Tomato is a vegetable crop, preceded only by potato in terms of production and global consumption (Dias, 2012). It is an important industrial and cash crop in many countries, because of the economically attractive and rich nutrient composition of the fruit, with the attendant health-associated benefits (Willcox et al., 2003). Tomato is being used in Nigeria as ingredients of meals, salads, ketchup, soups and sauces from time immemorial (Akaeze and Aduramigba-Modupe, 2017). Nigeria ranks 11th among the largest tomato producing countries in the world, with a production of 4.1 MT (FAOSTAT, 2017). However post-harvest loss of tomato is very alarming, estimated at 30%–50% in the country (Ajagbe et al., 2014). This is largely because the shelf life of the crop is particularly shortened (48 h) in the tropics (Muhammad et al., 2011), coupled with poor post-harvest handling and storage. To meet domestic demand for tomato, Nigeria imports 150000 MT of tomato paste annually, valued at 170 million USD (Ayedun and Akande, 2023). Moreover, a survey by Ugonna et al. (2015) revealed that only 20% of processed tomato are produced in Nigeria while the remaining 80% is imported. The fruits of Flavr-Savr™, a transgenic tomato variety released in 1994, was the first commercially available food crop. Since then, many other GM tomato varieties have been commercialized. Flavr-Savr™ was developed by inhibition of polygalacturonase enzyme, responsible for pectin molecule degradation in the cell wall, thereby causing delayed softening of fruit and elongated shelf life (Baranski et al., 2019). RNAi silencing and CRISPR-based mutation of ripening-related gene which encodes pectate lyase, caused the fruits to be firmer over a long period (Yang et al., 2017; Wang et al., 2019). Decrease in the activities of enzymes (polygalacturonase, tomato β-galactosidase, cellulase β-D-xylosidase) involved in cell wall modification as result of RNA silencing of SIFSR gene also elongated shelf-life significantly (Zhang et al., 2018). This makes SIFSR gene a modification a potential target for improving potato shelf-life. Similarly, overexpression of certain genes like SICOBRA-like gene, by genetic engineering revealed their role in elongating tomato shelf life (Cao et al., 2012).
The susceptibility of the current tomato cultivars to diseases and pests is another barrier to tomato production (Ugonna et al., 2015). Although, certain cultivars including H9-1-6 and Ronita are either resistant to leaf diseases or moderately resistant to root-knot nematode, they are susceptible to a host of others. The genetic basis of tomato is progressively becoming narrower from the time of domestication in its centres of origin to its spread to other parts of the world because selection was solely aimed at increasing yield (Gruber, 2017). Miller and Tanksley, (1990) reported that the genetic variation existing between tomato cultivars is Zhang et al., 2002; Rodríguez et al., 2006; Bai and Lindhout, 2007). Effective use of wild species of tomato for improving elite varieties requires accurate understanding of the genetic factors responsible for desirable agronomic traits in this wild species. However, most breeding projects are conventional in design, which makes simultaneous studying of multiple traits a difficult task. Since domestication of tomato from the wild species was almost solely driven by yield while other important traits such as resistance to diseases and tolerance to stress largely remained with the wild species (Kik et al., 2010), genome editing already proved useful in domesticating wild tomato (Zsögön et al., 2018). Using CRISPR-system technology, Zsögön et al. (2018), edited six loci—associated with high yield in elite tomato cultivars—in Solanum pimpinellifolium which resulted in alteration in the plant morphology alongside fruits size, number and higher accumulation of antioxidant lycopene.
Molecular farming
In 2021, Nigeria joined the rest of the world in the effort to develop an effective vaccine against the deadly coronavirus. Consequently, the Nigeria Vaccine Policy (NVP) was established for the first time in order to promote domestic vaccine manufacturing and guarantee autonomy in vaccine accessibility. A possible area of research that ought to be looked into is the use of plants in the production of effective vaccines against both communicable and non-communicable diseases. This is because the government offered to support and fund vaccine research and development as part of the implementation strategies to achieve the aim and objectives of the NVP. For over three decades, plants have been used as a bio-factory to manufacture pharmaceutically important recombinant proteins (such as plasma proteins, antibodies and cytokines) and diagnostic reagents through molecular farming (Fischer and Buyel, 2020). More recently, utilization of plants as subunit vaccine is gaining prominence, because it is a quicker and safer alternative to conventional vaccine development which is based on attenuation or inactivation of specific virus (Capell et al., 2020). The recombinant protein is produced in plant using deconstructed vector mediated by agroinfiltration with Agrobacterium tumefaciens (Gleba et al., 2014). Before the advent of molecular farming, there are established platforms such as microbes (e.g., Escherichia coli) and various mammalian cells cultures used in the industrial manufacturing of biologics. Plant is yet to displace these major platforms because investment in the industry and the existing regulatory framework favours these earlier established platforms. However, since Nigeria is just developing capacity in this area, government can take advantage of plants as the platform for the manufacturing of important biologics. This is because, with plants as the platform, the production of biologics can be done on a massive level and scaled up rapidly to cater for unexpected surge in demand and do not support growth of human pathogens (Ma et al., 2003; Whaley et al., 2011).
Origin of anti-GMO campaign in the world
In the early 1980s, scientists established Agrobacterium-mediated transformation in plants and identified CaMV 35S promoter, which can facilitate gene expression (Odell et al., 1985; Herrera-Estrella et al., 1992). These two important findings were later combined to engineer the production of the first transgenic herbicide tolerant plant (Shah et al., 1986). The three milestones led to the launch of plant biotechnology on a grand scale and created an avenue for the development of the field of recombinant DNA technology. Crops produced through this “unnatural method” of altering plant genetic material are regarded as genetically modified GM-crops (Abdul Aziz et al., 2022), although in the true sense, all crops, with respect to their current genetic make-up, originated from long time-controlled breeding, selection and domestication processes, which have genetically modified them from their wild state. However, regardless of the huge potential of GM-crops and their new possibilities in ensuring food security, there is a global skepticism about the consumption of such crops, because they are perceived to portend great risk to human health. For instance, heated debates among scientists on the safety of GM-crops came about from the findings of Arpad Pusztai, a protein scientist, who tested the effect of consumption of transgenic potato on rats, and afterwards, opined that consumption of such potato by the rats was the reason for the lack of good health of the animals (Enserink, 1998; 1999a). Even though no firm conclusion was made in his research to affirm the risk of GM-crops to human health, his work stared up anti-GM campaigns and fuelled further research. In the publications by (Ho et al. (1999) and the Institute for Agriculture and Trade Policy (1999), CaMV 35S promoter was regarded as the culprit in the supposed unwholesome effect of GM-crop, because it was assumed to be virus, whereas it is just a short stretch of DNA (Amack and Antunes, 2020). However, the concern raised by the Pusztai data was later debunked, after his data was re-examined by external experts and The Royal British Society, who found the framework of his experiment inconsistent and thereafter concluded that any finding from his work should be discarded as it lacked merit (Enserink, 1999b; Kuiper et al., 1999; The Royal Society, 1999; Yang, 2005). However in Nigeria, pressure groups made up of the coalition of civil society groups, farmers, students and faith-based organisations have constantly protested the adoption of GM-crops (Omeje, 2019). The argument against GM-crops is that it poses great threat in the areas of toxicology, allergy and immune dysfunction. These claims mostly originated from (Fagan et al., 2015), who stated that the process of genetic engineering could disrupt pristine proteins or metabolic pathways, which may result in the production of toxins or allergens in food. Predictive animal testing is a method that is commonly used to assess food allergies, both for genetically modified and non-GM foods. However, it has been found to be inadequate. (Wal, 2015). Till date, despite efforts toward developing animal models for accurate prediction of sensitivity to allergy, none has proven to be predictive (Ladics and Selgrade, 2009; Goodman, 2015; Kazemi et al., 2023). Consequent on this fact, the National Academies of Sciences, Engineering, and Medicine (2016), suggested the use of pre-commercialization tests to make rough predictions on relationships between consumption of GM-crops and the prevalence of some human diseases.
Plant biotechnology protest in Nigeria: the case study of transgenic cassava trial
Nigeria is yet to reach its full potential in cassava production despite being the leading producer. The main factor responsible for this is post-harvest loss due to rapid physiological deterioration of its swollen storage roots that are rich in starch. This reduces the post-harvest value to about 40% lower than its original worth and impact farmers’ income negatively. Addressing this problem will positively impact Nigeria in food security and boost socio-economic status of many farmers. Using techniques in plant biotechnology, the lab of Prof. Samuel C. Zeeman developed transgenic cassava called AMY3 RNAi transgenic lines. It is so named because the transgenic line lacks the activity of one of alpha-amylase isoforms AMY3, an enzyme involved in starch degradation in its storage roots and the named lines showed prospect of slowing down post-harvest deterioration of cassava storage root. In 2017, application for confined field trial of AMY3 RNAi transgenic lines at International Institute of Tropical Agriculture was approved by NBMA. Going forward, objections were raised against this, stating that the concerned lab has worked mostly on Arabidopsis previously (Fulton et al., 2008; Liu et al., 2023). However, it is a known fact that the evolution of starch metabolism is primarily conserved across angiosperm, which implies that knowledge gained from studying starch metabolism in the model plant, is mostly applicable to other plant species (Pfister and Zeeman, 2016). Although slight differences have been reported on biosynthesis and degradation of starch in storage organs like tuber and endosperm, Prof. Zeeman’s lab has demonstrated profound knowledge on starch metabolism in crops like cassava (Zhou et al., 2017; Bull et al., 2018; Wang et al., 2018) and potato (Hussain et al., 2003; Ferreira et al., 2017; Samodien et al., 2018) with several published reviews on this subject (Smith et al., 2005; Zeeman et al., 2010; Santelia and Zeeman, 2011; Stitt and Zeeman, 2012; Streb and Zeeman, 2012; Pfister and Zeeman, 2016; Smith and Zeeman, 2020). Thus, it is believed that he has wealth of experience to achieve this feat and the outcome of the research will be credible. Also, the claim that the work was not peer viewed before it was presented for trial in Nigeria may be a little bit premature at the time due to the fact that the transgenic cassava had been tested in greenhouse for 3 years in ETHZ Biotechnology Lab in Zurich, Switzerland. As a result, it makes sense to test the transgenic cassava’s performance in a natural cassava growing environment, and the application was approved through the proper channels. Additionally, not all scientific discovery is peer-reviewed before patenting. The Nigerian government’s takeaway from this experience ought to be to fortify the nation’s scientific workforce or promote joint research endeavours between Nigerian scientists and their international counterparts in order to progressively employ plant biotechnology instruments to tackle issues concerning food security in the nation.
Perspective and conclusion
In Nigeria, many people are involved in the direct production of various food crops. In fact, the country currently leads in the production of certain food crops globally, but this is usually due to increase in land conversion to crop production and not because of increase in yield/hectare. Production of other food crops is also suboptimal due to limiting factors like disease-infested planting material and susceptibility to biotic and abiotic stresses. While progress has been made using conventional breeding methods to develop crop varieties that are resistant, tolerant, or well adapted to both biotic and abiotic stress, this method is still limited in some ways. Techniques of plant biotechnology, such as genetic engineering and genome editing are increasingly becoming viable and sustainable options for improving our crops under certain conditions or circumstances. This review, therefore, provides general overview of the status of biotechnology in Africa with specific focus on Nigeria. Areas where biotechnological techniques will be most needed for crop improvements were identified and scientists can begin to take advantage of these novel techniques to provide lasting solutions to major problems, preventing Nigeria from reaching its full potential in crop production. Presently, the Nigerian government has enacted policies and regulations that favour responsible application and development of biotechnological products according to international best practice. Nigeria seems to be championing commercialization of biotech crops in Africa with recent approval of more genetically modified crops. Therefore, we recommend that governments in other African countries should take a cue from Nigeria by setting the stage for their countries to benefit from this technology towards ensuring food security and economic prosperity on the continent. Additionally, Nigerian government and start-up companies should also tap into immense potential of molecular farming for developing and production of subunit vaccines to bring about revenue and employment generation. The opposition from pressure groups against biotechnological application in Nigeria is largely due to the myth portraying the technology as foreign and not based on empirical evidences. For over two decades, several countries have benefited from sales and consumption of biotechnology products without any negative effects on the health of consumers.
Author contributions
MA: Conceptualization, Writing–original draft, Writing–review and editing, Investigation, Methodology. TA: Writing–review and editing, Investigation. IA: Writing–review and editing, Investigation. TO: Writing–review and editing. SA-S: Writing–review and editing. OF: Writing–review and editing. OS: Writing–original draft, Writing–review and editing, Supervision, Validation. CA: Supervision, Validation, Writing–original draft, Writing–review and editing. OA: Conceptualization, Supervision, Validation, Writing–original draft, Writing–review and editing, Methodology.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdul Aziz, M., Brini, F., Rouached, H., and Masmoudi, K. (2022). Genetically engineered crops for sustainably enhanced food production systems. Front. Plant Sci. 13, 1027828. doi:10.3389/fpls.2022.1027828
PubMed Abstract | CrossRef Full Text | Google Scholar
Abiwon, B. O., Dambaba, N., and Salihu, B. Z. (2016). Genetic improvement of rice in Nigeria for enhanced yeild and grain quality – a Review. Asian Res. J. Agric. 3, 1–18. doi:10.9734/ARJA/2016/28675
CrossRef Full Text | Google Scholar
Abrecht, D. G., and Carberry, P. S. (1993). The influence of water deficit prior to tassel initiation on maize growth, development and yield. Field Crops Res. 31, 55–69. doi:10.1016/0378-4290(93)90050-W
CrossRef Full Text | Google Scholar
Acheampong, P., Osei-adu, J., Amengo, E., and Sagoe, R. (2015). Cocoyam value chain and benchmark study in Ghana. doi:10.13140/rg.2.1.4295.6326
CrossRef Full Text | Google Scholar
Adejumo, B. A., Okundare, B., and Balogun, S. A. (2013). Quality attributes of yam flour (Elubo) as affected by blanching water temperature and soaking time. Int. J. Eng. Sci. 2, 216–221.
CrossRef Full Text | Google Scholar
Adekiya, A. O., and Agbede, T. M. (2016). The influence of three years of tillage and poultry manure application on soil and leaf nutrient status, growth and yield of cocoyam. J. Adv. Agric. Technol. 3, 104–109. doi:10.18178/joaat.3.2.104-109
CrossRef Full Text | Google Scholar
Africa Common Position on Food Systems Food Security Leadership Dialogue (ACPOFS) (2021). Regional submission to UN food systems summit. Retrieved from african common position on food systems | AUDA-NEPAD.
Google Scholar
Africa Harvest Biotech Foundation International (AHBFI) (2023). Agricultural biotechnology market size. Report 2023-2032 (precedenceresearch.com).
Google Scholar
Agnoun, Y., Biaou, S. S. H., Sié, M., Vodouhè, R. S., and Ahanchede, A. (2012). The african rice Oryza glaberrima steud: knowledge distribution and prospects. IJB 4, 158. doi:10.5539/ijb.v4n3p158
CrossRef Full Text | Google Scholar
Agueguia, A., and Fatokun, C. A. (1987). Pollen storage in cocoyam (Xanthosoma sagittifolium [L.] Schott). Euphytica 39, 195–198. doi:10.1007/bf00037095
CrossRef Full Text | Google Scholar
Aher, R. R., Reddy, P. S., Bhunia, R. K., Flyckt, K. S., Shankhapal, A. R., Ojha, R., et al. (2022). Loss-of-function of triacylglycerol lipases are associated with low flour rancidity in pearl millet [Pennisetum glaucum (L.) R. Br.]. Front. Plant Sci. 13, 962667. doi:10.3389/fpls.2022.962667
PubMed Abstract | CrossRef Full Text | Google Scholar
Aidoo, R., Nimoh, F., Bakang, J. E. A., Ohene-Yankyera, K., Fialor, S. C., and Abaidoo, R. C. (2011). Economics of small-scale seed yam production in Ghana: implications for commercialization. J. Sustain. Dev. Afr. 13, 65–78.
Google Scholar
Aighewi, B. A., Asiedu, R., Maroya, N., and Balogun, M. (2015). Improved propagation methods to raise the productivity of yam (Dioscorea rotundata Poir.). Food Sec 7, 823–834. doi:10.1007/s12571-015-0481-6
CrossRef Full Text | Google Scholar
Ajagbe, B. O., Oyediran, W. O., Omoare, A. M., and Sofowora, O. O. (2014). Assessment of post-harvest practices among tomato (Solanum lycopersicum) farmers/processors in Abeokuta North local government area of Ogun state, Nigeria. Int. J. Educ. Res. 2, 1–12.
Google Scholar
Ajeesh Krishna, T. P., Maharajan, T., and Ceasar, S. A. (2022). Improvement of millets in the post-genomic era. Physiology Mol. Biol. plants Int. J. Funct. plant Biol. 28 (3), 669–685. doi:10.1007/s12298-022-01158-8
CrossRef Full Text | Google Scholar
Ajeigbe, H. A., Waliyar, F., Echekwu, C. A., Ayuba, K., Motagi, B. N., Eniayeju, D., et al. (2014) “A farmer’s guide to groundnut production in Nigeria,” in Patancheru 502 324. Telangana, India: International Crops Research Institute for the Semi-Arid Tropics, 36.
Google Scholar
Akaeze, O., and Aduramigba-Modupe, A. O. (2017). Fusarium wilt disease of tomato: screening for resistance and in-vitro evaluation of botanicals for control; the Nigeria case. JMBFS 9, 32–36. doi:10.15414/jmbfs.2017.7.1.32-36
CrossRef Full Text | Google Scholar
Akano, A. O., Dixon, A. G. O., Mba, C., Barrera, E., and Fregene, M. (2002). Genetic mapping of a dominant gene conferring resistance to cassava mosaic disease. Theor. Appl. Genet. 105, 521–525. doi:10.1007/s00122-002-0891-7
PubMed Abstract | CrossRef Full Text | Google Scholar
Akinbo, O., Gedil, M., Ekpo, E. J. A., Oladele, J., and Dixon, A. G. O. (2007). Detection of RAPD markers-linked to resistance to cassava anthracnose disease. Afr. J. Biotechnol. 6, 677–682.
Google Scholar
Akinbo, O., Labuschagne, M., and Fregene, M. (2011). Introgression of whitefly (Aleurotrachelus socialis) resistance gene from F1 inter-specific hybrids into commercial cassava. Euphytica 183, 19–26. doi:10.1007/s10681-011-0436-8
CrossRef Full Text | Google Scholar
Akinbo, O., Labuschagne, M., and Fregene, M. (2012). Increased storage protein from interspecific F1 hybrids between cassava (Manihot esculenta Crantz) and its wild progenitor (M. esculenta ssp. flabellifolia). Euphytica 185, 303–311. doi:10.1007/s10681-011-0590-z
CrossRef Full Text | Google Scholar
Akinbo, O., Labuschagne, M., Marín, J., Ospina, C., Santos, L., Barrera, E., et al. (2012). QTL analysis for root protein in a backcross family of cassava derived from Manihot esculenta ssp flabellifolia. Trop. Plant Biol. 5, 161–172. doi:10.1007/s12042-012-9095-8
CrossRef Full Text | Google Scholar
Alliance for science (2024). Availble online at: Nigeria approves commercial release of GM maize varieties – alliance for Science (Accessed on February 16, 2024).
Google Scholar
Al-Shafeay, A. F., Ibrahim, A. S., Nesiem, M. R., and Tawfik, M. S. (2011). Establishment of regeneration and transformation system in Egyptian sesame (Sesamum indicum L.) cv Sohag 1. Gm. Crops 2 (3), 182–192. doi:10.4161/gmcr.2.3.18378
PubMed Abstract | CrossRef Full Text | Google Scholar
Alwang, J., Norton, G., and Larochelle, C. (2019). Obstacles to widespread diffusion of IPM in developing countries: lessons from the field. JIPM 10 (1), 10. doi:10.1093/jipm/pmz008
CrossRef Full Text | Google Scholar
Amack, S. C., and Antunes, M. S. (2020). CaMV35S promoter – a plant biology and biotechnology workhorse in the era of synthetic biology. Curr. Plant Biol. 24, 100179. doi:10.1016/j.cpb.2020.100179
CrossRef Full Text | Google Scholar
Amusa, N. A., Adegbite, A. A., Muhammed, S., and Baiyewu, R. A. (2003). Yam diseases and its management in Nigeria. Afr. J. Biotechnol. 2 (12), 497–502. doi:10.5897/ajb2003.000-1099
CrossRef Full Text | Google Scholar
Anilakumar, K. R., Pal, A., Khanum, F., and Bawa, A. S. (2010). Nutritional, medicinal and industrial uses of sesame (Sesamum indicum L.) seeds – an overview. Agric. Conspec. Sci. 75, 159–168.
Google Scholar
Antonio, G. C., Takeiti, C. Y., De Oliveira, R. A., and Park, K. J. (2011). Sweet potato: production, Morphological and Physicochemical characteristics, and technological process. Fruit, Veg. Cereal Sci. Biotechnol. 5, 1–18.
Google Scholar
Arcadia Biosciences, (2018). Final Report Available at: PA00TM4V.pdf (usaid.gov).
Google Scholar
Asala, S., Alegbejo, M. D., Kashina, B., Banwo, O. O., Asiedu, R., and Lava-Kumar, P. (2012). Distribution and incidence of viruses infecting yam (Dioscorea spp.) in Nigeria. GJBB 1, 163–167.
Google Scholar
Asante, S. K., Mensah, G. W. K., and Wahaga, E. (2008). Farmers’ knowledge and perceptions of insect pests of yam (Dioscorea spp.) and their indigenous control practices in Northern Ghana. Ghana Jnl Agric. Sci. 40, 185–192. doi:10.4314/gjas.v40i2.2169
CrossRef Full Text | Google Scholar
Avramovic, M. (1996). An affordable development?: biotechnology, economics, and the implications for the third world. London: Zed Books.
Google Scholar
Awata, L. A. O., Ifie, B. E., Danquah, E., Jumbo, M. B., Suresh, L. M., Gowda, M., et al. (2021). Introgression of Maize Lethal Necrosis Resistance quantitative trait loci into susceptible maize populations and validation of the resistance under field conditions in Naivasha, Kenya. Front. Plant Sci. 12, 649308. doi:10.3389/fpls.2021.649308
PubMed Abstract | CrossRef Full Text | Google Scholar
Ayedun, B., and Akande, A. (2023). Socioeconomic Effects of Oyo State government COVID-19 palliatives on tomato smallholder farmers. Int. J. Agric. Veterinary Sci. 5, 52–63. doi:10.34104/ijavs.023.052063
CrossRef Full Text | Google Scholar
Baah, F. D., Maziya-Dixon, B., Asiedu, R., Oduro, I., and Ellis, W. O. (2009). Nutritional and biochemical composition of D. alata (Dioscorea spp.) tubers. J. Food Agric. Environ. 7, 373–378.
Google Scholar
Baranski, R., Klimek-Chodacka, M., and Lukasiewicz, A. (2019). Approved genetically modified (GM) horticultural plants: a 25-year perspective. Folia Hortic. 31, 3–49. doi:10.2478/fhort-2019-0001
CrossRef Full Text | Google Scholar
Beyene, G., Solomon, F. R., Chauhan, R. D., Gaitán-Solis, E., Narayanan, N., Gehan, J., et al. (2018). Provitamin A biofortification of cassava enhances shelf life but reduces dry matter content of storage roots due to altered carbon partitioning into starch. Plant Biotechnol. J. 16, 1186–1200. doi:10.1111/pbi.12862
PubMed Abstract | CrossRef Full Text | Google Scholar
Bharose, A. A., Hajare, S. T., Narayanrao, D. R., Gajera, H. G., Prajapati, H. K., Singh, S. C., et al. (2024). Whole genome sequencing and annotation of Aspergillus flavus JAM-JKB-B HA-GG20. Sci. Rep. 14 (18), 18. doi:10.1038/s41598-023-50986-5
PubMed Abstract | CrossRef Full Text | Google Scholar
Bhatnagar-Mathur, P., Sunkara, S., Bhatnagar-Panwar, M., Waliyar, F., and Sharma, K. K. (2015). Biotechnological advances for combating Aspergillus flavus and aflatoxin contamination in crops. Plant Sci. 234, 119–132. doi:10.1016/j.plantsci.2015.02.009
PubMed Abstract | CrossRef Full Text | Google Scholar
Bhatt, R., Asopa, P. P., Jain, R., Kothari-Chajer, A., Kothari, S. L., and Kachhwaha, S. (2021). Optimization of Agrobacterium mediated genetic transformation in Paspalum scrobiculatum L. (Kodo millet). Agronomy 11, 1104. doi:10.3390/agronomy11061104
CrossRef Full Text | Google Scholar
Blümmel, M., Anandan, S., and Wright, I. A. (2012). “Improvement of feed resources and livestock feeding in mixed cropping systems,” in Animal nutrition advances and development. Editors U. R. Mehra, P. Singh, and A. K. Verma (Delhi, India: Satish Serial Publishing House), 459–475.
Google Scholar
Boch, J., Scholze, H., Schornack, S., Landgraf, A., Hahn, S., Kay, S., et al. (2009). Breaking the code of DNA binding specificity of TAL-Type III Effectors. Science 326, 1509–1512. doi:10.1126/science.1178811
PubMed Abstract | CrossRef Full Text | Google Scholar
Brar, G. S., and Ahuja, K. L. (2008). Sesame: its culture, genetics, breeding and biochemistry. Annu. Rev. Plant Sci. 1, 245–313.
Google Scholar
Brown, D. (2000). Aroids. “Plants of the arum family”. 2nd edn. Portland, OR, USA: Timber Press.
Google Scholar
Brummell, D. A., Watson, L. M., Zhou, J., McKenzie, M. J., Hallett, I. C., Simmons, L., et al. (2015). Overexpression of STARCH BRANCHING ENZYME II increases short-chain branching of amylopectin and alters the physicochemical properties of starch from potato tuber. BMC Biotechnol. 15, 28. doi:10.1186/s12896-015-0143-y
PubMed Abstract | CrossRef Full Text | Google Scholar
Bull, S. E., Seung, D., Chanez, C., Mehta, D., Kuon, J. E., Truernit, E., et al. (2018). Accelerated ex situ breeding of GBSS- and PTST1-edited cassava for modified starch. Sci. Adv. 4, eaat6086. doi:10.1126/sciadv.aat6086
PubMed Abstract | CrossRef Full Text | Google Scholar
Businessday NG (2023). Nigeria’s 60% post-harvest loss creates investment opportunity for investors – Businessday NG.
Google Scholar
Cao, Y., Tang, X., Giovannoni, J., Xiao, F., and Liu, Y. (2012). Functional characterization of a tomato COBRA-like gene functioning in fruit development and ripening. BMC Plant Biol. 12, 211. doi:10.1186/1471-2229-12-211
PubMed Abstract | CrossRef Full Text | Google Scholar
Capell, T., Twyman, R. M., Armario-Najera, V., Ma, J. K. C., Schillberg, S., and Christou, P. (2020). Potential applications of plant biotechnology against SARS-CoV-2. Trends Plant Sci. 25, 635–643. doi:10.1016/j.tplants.2020.04.009
PubMed Abstract | CrossRef Full Text | Google Scholar
Ceasar, S., and Ignacimuthu, S. (2011). Agrobacterium-mediated transformation of finger millet (Eleusine coracana (L.) Gaertn.) using shoot apex explants. Plant Cell. Rep. 30, 1759–1770. doi:10.1007/s00299-011-1084-0
PubMed Abstract | CrossRef Full Text | Google Scholar
CGIAR (2022). Genome editing for tolerance to maize lethal necrosis – cgiar (accessed on October 01, 2024).
Google Scholar
Chen, Y., Sun, X., Zhou, X., Hebelstrup, K. H., Blennow, A., and Bao, J. (2017). Highly phosphorylated functionalized rice starch produced by transgenic rice expressing the potato GWD1 gene. Sci. Rep. 7, 3339. doi:10.1038/s41598-017-03637-5
PubMed Abstract | CrossRef Full Text | Google Scholar
Cheng, Z., Sun, Y., Yang, S., Zhi, H., Yin, T., Ma, X., et al. (2021). Establishing in planta haploid inducer line by edited SiMTL in foxtail millet (Setaria italica). Plant Biotechnol. J. 19, 1089–1091. doi:10.1111/pbi.13584
PubMed Abstract | CrossRef Full Text | Google Scholar
Chevalier, B. S., and Stoddard, B. L. (2001). Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res. 29, 3757–3774. doi:10.1093/nar/29.18.3757
PubMed Abstract | CrossRef Full Text | Google Scholar
Chowdhury, S., Basu, A., and Kundu, S. (2014). A new high-frequency Agrobacterium-mediated transformation technique for Sesamum indicum L. using de-embryonated cotyledon as explant. Protoplasma 251, 1175–1190. doi:10.1007/s00709-014-0625-0
PubMed Abstract | CrossRef Full Text | Google Scholar
Chowdhury, S., Basu, A., and Kundu, S. (2017). Overexpression of a new Osmotin-Like Protein Gene (SindOLP) confers tolerance against biotic and abiotic stresses in Sesame. Front. Plant Sci. 8, 410. doi:10.3389/fpls.2017.00410
PubMed Abstract | CrossRef Full Text | Google Scholar
Cole, R. J., Dorner, J. W., and Holbrook, C. C. (1995). “Advances in mycotoxin elimination and resistance,” in Advances in peanut science. Amer. Peanut res. And educ. Soc. Editors H. E. Pattee, and H. T. Stalker (United States: Stillwater), 456–474.
Google Scholar
CSC news (2024) Cuba offers the world healthcare alternatives. cuba-solidarity.org.uk) (Accessed February 16, 2024).
Google Scholar
CSSP (2023). Cassava source-sink project. (cass-research.org).
Google Scholar
Datta, S. K. (2004). Rice biotechnology: a need for developing countries. AgBioForum 7, 31–35.
Google Scholar
Degefa, I., and Anbessa, B. (2017). Agricultural practices and traditional preservation of taro (Colocasia spp.) in abaya woreda, southern Ethiopia. Adv. Biotechnol. Microbiol. 7, 1–7. doi:10.19080/AIBM.2017.07.555723
CrossRef Full Text | Google Scholar
de Lourdes Contreras-Pacheco, M., Santacruz-Ruvalcaba, F., García-Fajardo, J. A., de Jesús Sánchez, G. J., Ruíz, L., Estarrón-Espinosa, M., et al. (2013). Diosgenin quantification, characterisation and chemical composition in a tuber collection of Dioscorea spp. in the state of Jalisco, Mexico. Int. J. Food Sci. Technol. 48, 2111–2118. doi:10.1111/ijfs.12193
CrossRef Full Text | Google Scholar
Dias, J. S. (2012). Nutritional quality and health benefits of vegetables: a review. Food Nutr. Sci. 3, 1354–1374. doi:10.4236/fns.2012.310179
CrossRef Full Text | Google Scholar
Dossa, K., Diouf, D., Wang, L., Wei, X., Zhang, Y., Niang, M., et al. (2017). The emerging oilseed crop Sesamum indicum enters the “Omics” era. Front. Plant Sci. 8, 1154. doi:10.3389/fpls.2017.01154
PubMed Abstract | CrossRef Full Text | Google Scholar
Dugassa, G., and Feyissa, T. (2011). In vitro production of virus-free sweet potato (Ipomoea batatas (l.) Lam) by meristem culture and thermotherapy. Ethiop. J. Sci. 34, 17–28. doi:10.4314/SINET.V34I1.78226
CrossRef Full Text | Google Scholar
Dutta, S., Muthusamy, V., Zunjare, R. U., and Hossain, F. (2019). Analysis of paralogous genes of Carotenoid dioxygenase affecting carotenoid biosynthesis pathway in maize (Zea mays L.). J. Pharmacogn. Phytochem. 8, 524–530.
Google Scholar
Egeonu, I. N., and Akoroda, M. O. (2010). Sweet potato characterization in Nigeria. Sweet potato Breeders’ Annu. Meet. Mukono, Uganda, 1–31.
Google Scholar
Elangovan, M., Venkatesh, K., Pandey, S., and Pandey, C. D. (2022). International Year of Millets 2023: opportunity for enhancing the use of Indian millets germplasm. Indian J. Plant Genet. Resour. 35, 90–94. doi:10.5958/0976-1926.2022.00048.1
CrossRef Full Text | Google Scholar
Ewa, F. (2021). Genetic mapping and evaluation of cassava (manihot esculenta crantz) for drought tolerance and early bulking in marginal Savannah ecology of Nigeria (Doctoral dissertation).
Google Scholar
Federal Ministry of Health Nigeria (FMHN) Department of community development and population activities nutrition division the Nigeria food consumption and nutrition survey (2005). National guidelines on micronutrients deficiencies control in Nigeria. Nigeria: Abuja.
Google Scholar
Ferreira, S. J., Senning, M., Fischer-Stettler, M., Streb, S., Ast, M., Neuhaus, H. E., et al. (2017). Simultaneous silencing of isoamylases ISA1, ISA2 and ISA3 by multi-target RNAi in potato tubers leads to decreased starch content and an early sprouting phenotype. PLoS ONE 12, e0181444. doi:10.1371/journal.pone.0181444
PubMed Abstract | CrossRef Full Text | Google Scholar
Fosu-Nyarko, J., and Jones, M. G. K. (2015). “Application of biotechnology for nematode control in crop plants,” in Plant nematode interactions. Editors C. Escobar, C. B. T.-A, and B. R. Fenoll (Academic Press), 73, 339–376. doi:10.1016/bs.abr.2014.12.012
CrossRef Full Text | Google Scholar
Fountain, J. C., Clevenger, J. P., Nadon, B., Youngblood, R. C., Korani, W., Chang, P. K., et al. (2020). Two new Aspergillus flavus reference genomes reveal a large insertion potentially contributing to isolate stress tolerance and aflatoxin production. G3 (Bethesda, Md.) 10 (10), 3515–3531. doi:10.1534/g3.120.401405
PubMed Abstract | CrossRef Full Text | Google Scholar
Fregene, M., Angel, F., Gomez, R., Rodriguez, F., Chavarriaga, P., Roca, W., et al. (1997). A molecular genetic map of cassava (Manihot esculenta Crantz). Theor. Appl. Genet. (1997) 95, 431–441. doi:10.1007/s001220050580
CrossRef Full Text | Google Scholar
Fregene, M., Bernal, A., Duque, M., Dixon, A., and Tohme, J. (2000). AFLP analysis of African cassava (Manihot esculenta Crantz) germplasm resistant to the cassava mosaic disease (CMD). Theor. Appl. Genet. 100, 678–685. doi:10.1007/s001220051339
CrossRef Full Text | Google Scholar
Fregene, M., Morante, N., Sanchez, T., Marin, J., Ospina, C., Barrera, E., et al. (2006). Molecular markers for introgression of useful traits from wild Manihot relatives of cassava, marker assisted selection (MAS) of diseases and root quality traits. J. Root Crops 32, 1–31.
Google Scholar
Fritsche, S., Poovaiah, C., MacRae, E., and Thorlby, G. (2018). A New Zealand perspective on the application and regulation of gene editing. Front. Plant Sci. 12, 1323. doi:10.3389/fpls.2018.01323
PubMed Abstract | CrossRef Full Text | Google Scholar
Fufa, T. W., Oselebe, H. O., Amadi, C. O., Menamo, T. M., and Abtew, W. G. (2023). Genetic diversity and association of yield-related traits in Taro (Colocasia esculenta (L.) Schott) sourced from different agroecological origins of Nigeria. Int. J. Agron. 9, 1–9. doi:10.1155/2023/8832165
CrossRef Full Text | Google Scholar
Fukino, N., Hanada, K., Ajisaka, H., Sakai, J., Hirochika, H., Hirai, M., et al. (2000). Transformation of Taro (Colocasia esculenta Schott) using particle bombardment. Jpn. Int. Res. Cent. Agric. Sci. 34, 159–165.
Google Scholar
Fukuda, Y., Osawa, T., Namiki, M., and Ozaki, T. (1985). Studies on antioxidative substances in Sesame seed. Agric. Biol. Chem. 49, 301–306. doi:10.1271/bbb1961.49.301
CrossRef Full Text | Google Scholar
Fulton, D. C., Stettler, M., Mettler, T., Vaughan, C. K., Li, J., Francisco, P., et al. (2008). Beta-AMYLASE4, a noncatalytic protein required for starch breakdown, acts upstream of three active beta-amylases in Arabidopsis chloroplasts. Plant Cell. 20, 1040–1058. doi:10.1105/tpc.107.056507
PubMed Abstract | CrossRef Full Text | Google Scholar
Futakuchi, K., Fofana, M., and Sie, M. (2008). Varietal differences in lodging resistance of African rice (Oryza glaberrima Steud.). Asian J. Plant Sci. 7, 569–573. doi:10.3923/ajps.2008.569.573
CrossRef Full Text | Google Scholar
Gao, J., Zhang, T., Xu, B., Jia, L., Xiao, B., Liu, H., et al. (2018). CRISPR/Cas9-mediated mutagenesis of carotenoid cleavage dioxygenase 8 (CCD8) in tobacco affects shoot and root architecture. Int. J. Mol. Sci. 19, 1062. doi:10.3390/ijms19041062
PubMed Abstract | CrossRef Full Text | Google Scholar
Gayin, J., Abdel-Aal, E.-S. M., Manful, J., Bertoft, E., Marcone, M., and Ragaee, S. (2017). Physical, cooking and thermal properties of African rice (Oryza glaberrima) and its starch digestibility in vitro. LWT 75, 481–487. doi:10.1016/j.lwt.2016.09.023
CrossRef Full Text | Google Scholar
Gobena, D., Shimels, M., Rich, P. J., Ruyter-Spira, C., Bouwmeester, H., Kanuganti, S., et al. (2017). Mutation in sorghum LOW GERMINATION STIMULANT 1 alters strigolactones and causes Striga resistance. Proc. Natl. Acad. Sci. 114, 4471–4476. doi:10.1073/pnas.1618965114
PubMed Abstract | CrossRef Full Text | Google Scholar
Goergen, G., Kumar, P. L., Sankung, S. B., Togola, A., and Tamò, M. (2016). First report of outbreaks of the fall armyworm Spodoptera frugiperda (J. E Smith) (Lepidoptera, Noctuidae), a new alien invasive pest in West and Central Africa. PLOS ONE 11 (10), e0165632. doi:10.1371/journal.pone.0165632
PubMed Abstract | CrossRef Full Text | Google Scholar
Gomez, M. A., Lin, Z. D., Moll, T., Chauhan, R. D., Hayden, L., Renninger, K., et al. (2019). Simultaneous CRISPR/Cas9-mediated editing of cassava eIF 4E isoforms nCBP-1 and nCBP-2 reduces cassava brown streak disease symptom severity and incidence. Plant Biotechnol. J. 17 (2), 421–434. doi:10.1111/pbi.12987
PubMed Abstract | CrossRef Full Text | Google Scholar
Gomez-Roldan, V., Fermas, S., Brewer, P. B., Puech-Pagès, V., Dun, E. A., Pillot, J. P., et al. (2008). Strigolactone inhibition of shoot branching. Nature 455, 189–194. doi:10.1038/nature07271
PubMed Abstract | CrossRef Full Text | Google Scholar
Goodman, R. (2015). “Evaluating GE food sources for risks of allergy: methods, gaps and perspective,” in Presentation to the National Academy of Sciences’ Committee on genetically engineered crops: past experience and future Prospects.
Google Scholar
Goswami, S., Asrani, P., Ali, T. A., Kumar, R. D., Vinutha, T., Veda, K., et al. (2020). Rancidity matrix: development of biochemical indicators for analysing the keeping quality of pearl millet flour. Food Anal. Methods 13, 2147–2164. doi:10.1007/s12161-020-01831-2
CrossRef Full Text | Google Scholar
Hao, J., Yang, Y., Futrell, S., Kelly, E. A., Lorts, C. M., Nebie, B., et al. (2023). CRISPR/Cas9-mediated mutagenesis of carotenoid cleavage dioxygenase (CCD) genes in sorghum alters strigolactone biosynthesis and plant biotic interactions. Phytobiomes J. 7, 339–351. doi:10.1094/PBIOMES-08-22-0053-R
CrossRef Full Text | Google Scholar
Hariprasanna, K., Agte, V., Elangovan, M., and Patil, J. (2014). Genetic variability for grain iron and zinc content in cultivars, breeding lines and selected germplasm accessions of sorghum [Sorghum bicolor (L.) Moench]. Indian J. Genet. Plant Breed. 74, 42–49. doi:10.5958/j.0975-6906.74.1.006
CrossRef Full Text | Google Scholar
Harris-Fry, H., Nur, H., Shankar, B., Zanello, G., Srinivasan, C., and Kadiyala, S. (2020). The impact of gender equity in agriculture on nutritional status, diets, and household food security: a mixed-methods systematic review. BMJ Glob. health 5, e002173. doi:10.1136/bmjgh-2019-002173
PubMed Abstract | CrossRef Full Text | Google Scholar
Hatakeyama, M., Aluri, S., Balachadran, M. T., Sivarajan, S. R., Patrignani, A., Grüter, S., et al. (2017). Multiple hybrid de novo genome assembly of finger millet, an orphan allotetraploid crop. DNA Res. 25, 39–47. doi:10.1093/dnares/dsx036
PubMed Abstract | CrossRef Full Text | Google Scholar
He, X., Miyasaka, S. C., Fitch, M. M. M., Khuri, S., and Zhu, Y. J. (2013). Taro (Colocasia esculenta), transformed with a wheat oxalate oxidase gene for improved resistance to taro pathogen Phytophthora colocasiae. HortScience Horts 48, 22–27. doi:10.21273/HORTSCI.48.1.22
CrossRef Full Text | Google Scholar
He, X., Miyasaka, S. C., Fitch, M. M. M., Moore, P. H., and Zhu, Y. J. (2008). Agrobacterium tumefaciens-mediated transformation of taro (Colocasia esculenta (L.) Schott) with a rice chitinase gene for improved tolerance to a fungal pathogen Sclerotium rolfsii. Plant Cell. Rep. 27, 903–909. doi:10.1007/s00299-008-0519-8
PubMed Abstract | CrossRef Full Text | Google Scholar
He, X., Miyasaka, S. C., Fitch, M. M. M., and Zhu, Y. J. (2015) Taro (Colocasia esculenta (L.) schott) BT – Agrobacterium protocols. New York: Springer, 97–108. doi:10.1007/978-1-4939-1658-0_9
CrossRef Full Text | Google Scholar
He, X., Miyasaka, S. C., Zou, Y., Fitch, M. M. M., and Zhu, Y. J. (2010). Regeneration and transformation of taro (Colocasia esculenta) with a rice chitinase gene enhances resistance to Sclerotium rolfsii. HortScience Horts 45, 1014–1020. doi:10.21273/HORTSCI.45.7.1014
CrossRef Full Text | Google Scholar
Helliot, B., Panis, B., Poumay, Y., Swennen, R., Lepoivre, P., and Frison, E. (2002). Cryopreservation for the elimination of cucumber mosaic and banana streak viruses from banana (Musa spp.). Plant Cell. Rep. 20, 1117–1122. doi:10.1007/s00299-002-0458-8
CrossRef Full Text | Google Scholar
Herrera-Estrella, L., Depicker, A., Van Montagu, M., and Schell, J. (1992). Expression of chimaeric genes transferred into plant cells using a Ti-plasmid-derived vector. Biotechnol. Read. Mass 24, 377–381.
PubMed Abstract | Google Scholar
Hittalmani, S., Mahesh, H., Shirke, M. D., Biradar, H., Uday, G., Aruna, Y., et al. (2017). Genome and transcriptome sequence of finger millet (Eleusine coracana (L.) Gaertn.) provides insights into drought tolerance and nutraceutical properties. BMC Genomics 18, 465. doi:10.1186/s12864-017-3850-z
PubMed Abstract | CrossRef Full Text | Google Scholar
Ho, M. W., Ryan, A., and Cummins, J. (1999). Cauliflower mosaic viral promoter – a recipe for disaster? Microb. Ecol. Health Dis. 11, 194–197. doi:10.1080/089106099435628
CrossRef Full Text | Google Scholar
Holbrook, C. C., Guo, B. Z., Wilson, D. M., and Timper, P. (2009). The U.S. breeding program to develop peanut with drought tolerance and reduced aflatoxin contamination. PeanutSci. 36, 50–53. doi:10.3146/at07-009.1
CrossRef Full Text | Google Scholar
Howeler, R., Lutaladio, N., and Thomas, G. (2013) Save and Grow Cassava: a guide to sustainable production intensification. Rome: FAO.
Google Scholar
Huang, J., Yang, Q., and Pu, H. (2018). “Slowly digestible starch,” in Functional starch and applications in food. Editor Z. Jin (Singapore: Springer), 27–61. doi:10.1007/978-981-13-1077-5_2
CrossRef Full Text | Google Scholar
Hummel, A. W., Chauhan, R. D., Cermak, T., Mutka, A. M., Vijayaraghavan, A., Boyher, A., et al. (2018). Allele exchange at the EPSPS locus confers glyphosate tolerance in cassava. Plant Biotechnol. J. 16 (7), 1275–1282. doi:10.1111/pbi.12868
PubMed Abstract | CrossRef Full Text | Google Scholar
Hummel, M., Talsma, E. F., Van der Honing, A., Gama, A. C., Van Vugt, D., Brouwer, I. D., et al. (2018). Sensory and cultural acceptability tradeoffs with nutritional content of biofortified orange-fleshed sweetpotato varieties among households with children in Malawi. PLOS ONE 13, e0204754. doi:10.1371/journal.pone.0204754
PubMed Abstract | CrossRef Full Text | Google Scholar
Hundleby, P., and Harwood, W. (2022). “Regulatory constraints and differences of genome-edited crops around the globe,” in Genome editing: current technology advances and applications for crop improvement. Editors S. H. Wani, and G. Hensel (Springer International Publishing), 319–341. doi:10.1007/978-3-031-08072-2_17
CrossRef Full Text | Google Scholar
Hussain, H., Mant, A., Seale, R., Zeeman, S., Hinchliffe, E., Edwards, A., et al. (2003). Three isoforms of isoamylase contribute different catalytic properties for the debranching of potato glucans. Plant Cell. 15, 133–149. doi:10.1105/tpc.006635
PubMed Abstract | CrossRef Full Text | Google Scholar
Ignacimuthu, S., and Ceasar, S. A. (2012). Development of transgenic finger millet (Eleusine coracana (L.) Gaertn.) resistant to leaf blast disease. J. Biosci. 37, 135–147. doi:10.1007/s12038-011-9178-y
PubMed Abstract | CrossRef Full Text | Google Scholar
IITA (1989). International Institute of tropical Agriculture 1988/1989 annual report. Available at: U89SerIitaAnnualNothomNodev.pdf.
Google Scholar
Ikuemonisan, E. S., Mafimisebi, T. E., Ajibefun, I., and Adenegan, K. (2020). Cassava production in Nigeria: trends, instability and decomposition analysis (1970-2018). Heliyon 6, e05089. doi:10.1016/j.heliyon.2020.e05089
PubMed Abstract | CrossRef Full Text | Google Scholar
ISAAA (2023a). Mon87460| GM approval database- ISAAA.org.
Google Scholar
ISAAA, (2023b). Available at: Mon89034| GM approval database- ISAAA.org
Google Scholar
Ivancic, A., Garcia, J. Q., and Lebot, V. (2004). Genetically controlled branching corms of taro (Colocasia esculenta). N. Z. J. Crop Hortic. Sci. 32, 167–177. doi:10.1080/01140671.2004.9514293
CrossRef Full Text | Google Scholar
Janila, P., and Nigam, S. N. (2013). “Phenotyping for groundnut (Arachis hypogaea L.) improvement. Phenotyping for plant breeding,” in Phenotyping for plant breeding: applications of phenotyping methods for crop improvement. Editors S. K. Panguluri, and A. Ashok Kumar (New York: Springer), 129–167. doi:10.1007/978-1-4614-8320-5
CrossRef Full Text | Google Scholar
Janila, P., Nigam, S. N., Pandey, M. K., Nagesh, P., and Varshney, R. K. (2013). Groundnut improvement: use of genetic and genomic tools. Front. Plant Sci. 4 (4), 23. doi:10.3389/fpls.2013.00023
PubMed Abstract | CrossRef Full Text | Google Scholar
Jaouannet, M., Magliano, M., Arguel, M. J., Gourgues, M., Evangelisti, E., Abad, P., et al. (2013). The root-knot nematode calreticulin Mi-CRT is a key effector in plant defense suppression. Mol. Plant-Microbe Interact. MPMI 26, 97–105. doi:10.1094/MPMI-05-12-0130-R
PubMed Abstract | CrossRef Full Text | Google Scholar
Jenkins, M., Shanks, C. B., Brouwer, R., and Houghtaling, B. (2018). Factors affecting farmers’ willingness and ability to adopt and retain vitamin A-rich varieties of orange-fleshed sweet potato in Mozambique. Food Secur. 10, 1501–1519. doi:10.1007/s12571-018-0845-9
PubMed Abstract | CrossRef Full Text | Google Scholar
Jennings, D. (2009). “Tropical root and tuber crops. Cassava, sweet potato, yams and aroids,”. Editor V. Lebot (Wallingford, UK: CABI), 413. doi:10.1017/S0014479709007832
CrossRef Full Text | Google Scholar
Juma, B. S., Mukami, A., Mweu, C., Ngugi, M. P., and Mbinda, W. (2022). Targeted mutagenesis of the CYP79D1 gene via CRISPR/Cas9-mediated genome editing results in lower levels of cyanide in cassava. Front. Plant Sci. 13, 1009860. doi:10.3389/fpls.2022.1009860
PubMed Abstract | CrossRef Full Text | Google Scholar
Jung, K. H., An, G., and Ronald, P. C. (2008). Towards a better bowl of rice: assigning function to tens of thousands of rice genes. Nat. Rev. Genet. 9, 91–101. doi:10.1038/nrg2286
PubMed Abstract | CrossRef Full Text | Google Scholar
Kalu, C., Nnabue, I., Edemodu, A., Agre, P. A., Adebola, P., Asfaw, A., et al. (2023). Farmers’ perspective toward a demand led yam breeding in Nigeria. Front. Sustain. Food Syst. 7. doi:10.3389/fsufs.2023.1227920
CrossRef Full Text | Google Scholar
Karembu, M., and Ngure, G. (2022) Genome editing in Africa’s agriculture 2022: an early take-off. Nairobi Kenya: International Service for the Acquisition of Agri-biotech Applications.
Google Scholar
Kik, C., Visser, B., van Hintum, T., van Treuren, R., and van de Wouw, M. (2010). Genetic erosion in crops: concept, research results and challenges. Plant Genet. Resour. 8, 1–15. doi:10.1017/S1479262109990062
CrossRef Full Text | Google Scholar
Kim, Y. G., Cha, J., and Chandrasegaran, S. (1996). Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. U. S. A. 93, 1156–1160. doi:10.1073/pnas.93.3.1156
PubMed Abstract | CrossRef Full Text | Google Scholar
Kolte, S. J. (1985). Disease of annual edible oil seed crops. Vol II: rape seed-mustard and sesame diseases. Boca Raton: CRC Press.
Google Scholar
Kuiper, H. A., Noteborn, H. P., and Peijnenburg, A. A. (1999). Adequacy of methods for testing the safety of genetically modified foods. Lancet London, Engl. 354, 1315–1316. doi:10.1016/S0140-6736(99)00341-4
PubMed Abstract | CrossRef Full Text | Google Scholar
Kumar, A., Tomer, V., Kaur, A., Kumar, V., and Gupta, K. (2018). Millets: a solution to agrarian and nutritional challenges. Agric. Food Secur 7, 31. doi:10.1186/s40066-018-0183-3
CrossRef Full Text | Google Scholar
Kyetere, D., Okogbenin, E., Okeno, J., Sanni, K., Munyaradzi, J., Nangayo, F., et al. (2019). The role and contribution of plant breeding and plant biotechnology to sustainable agriculture in Africa. Afr. focus 32, 83–108. doi:10.1163/2031356x-03202008
CrossRef Full Text | Google Scholar
Ladics, G. S., and Selgrade, M. K. (2009). Identifying food proteins with allergenic potential: evolution of approaches to safety assessment and research to provide additional tools. Regul. Toxicol. Pharmacol. 54, S2–S6. doi:10.1016/j.yrtph.2008.10.010
PubMed Abstract | CrossRef Full Text | Google Scholar
León-de la O, D. I., Thorsteinsdóttir, H., and Calderón-Salinas, J. V. (2018). The rise of health biotechnology research in Latin America: a scientometric analysis of health biotechnology production and impact in Argentina, Brazil, Chile, Colombia, Cuba and Mexico. PLOS ONE 13, e0191267. doi:10.1371/journal.pone.0191267
PubMed Abstract | CrossRef Full Text | Google Scholar
Liang, Z., Wu, Y., Ma, L., Guo, Y., and Ran, Y. (2022). Efficient genome editing in Setaria italica using CRISPR/Cas9 and base editors. Front. Plant Sci. 12, 815946. doi:10.3389/fpls.2021.815946
PubMed Abstract | CrossRef Full Text | Google Scholar
Lin, C. S., Hsu, C. T., Yang, L. H., Lee, L. Y., Fu, J. Y., Cheng, Q. W., et al. (2018). Application of protoplast tech-nology to CRISPR/Cas9 mutagenesis: from single-cell mutation detection to mutant plant regeneration. Plant Biotechnol. J. 16, 1295–1310. doi:10.1111/pbi.12870
PubMed Abstract | CrossRef Full Text | Google Scholar
Liu, C., Pfister, B., Osman, R., Ritter, M., Heutinck, A., Sharma, M., et al. (2023). LIKE EARLY STARVATION 1 and EARLY STARVATION 1 promote and stabilize amylopectin phase transition in starch biosynthesis. Sci. Adv. 9 (21), eadg7448. doi:10.1126/sciadv.adg7448
PubMed Abstract | CrossRef Full Text | Google Scholar
Lloyd, J. R., Wilhelm, R., Sharma, M. K., Kossmann, J., and Zhang, P. (2023). Editorial: insights in plant biotechnology: 2021. Front. Plant Sci. 14, 1147930. doi:10.3389/fpls.2023.1147930
PubMed Abstract | CrossRef Full Text | Google Scholar
Loebenstein, G. (2009). in Origin, distribution and economic importance BT – the sweetpotato. Editors G. Loebenstein, and G. Thottappilly (Netherlands: Springer), 9–12. doi:10.1007/978-1-4020-9475-0_2
CrossRef Full Text | Google Scholar
Long, C., and Liu, K. (2001). Colocasia lihengiae (araceae: colocasieae), a new species from yunnan, China. Botanical Bull. Acad. Sinica 42, 313–317.
Google Scholar
Martin, D. K., Vicente, O., Beccari, T., Kellermayer, M., Koller, M., Lal, R., et al. (2021). A brief overview of global biotechnology. Biotechnol. Biotechnol. Equip. 35, S5–S14. doi:10.1080/13102818.2021.1878933
CrossRef Full Text | Google Scholar
Mattes, R. D., Kris-Etherton, P. M., and Foster, G. D. (2008). Impact of peanuts and tree nuts on body weight and healthy weight loss in adults. J. Nutr. 138, 1741–1745. doi:10.1093/jn/138.9.1741S
PubMed Abstract | CrossRef Full Text | Google Scholar
M’Boob, S. S. (1989). “A regional program for striga control in West and central africa,” in Striga – improved management in Africa. FAO plant production and protection paper. Editors T. O. Robson, and H. R. Broad (Rome: Food and Agriculture Organization), 96, 190–194.
Google Scholar
McCallum, E. J., Anjanappa, R. B., and Gruissem, W. (2017). Tackling agriculturally relevant diseases in the staple crop cassava (Manihot esculenta). Curr. Opin. Plant Biol. 38, 50–58. doi:10.1016/j.pbi.2017.04.008
PubMed Abstract | CrossRef Full Text | Google Scholar
McNeil, B. C., and Dean, D. H. (2011). Bacillus thuringiensis Cry2Ab is active on Anopheles mosquitoes: single D block exchanges reveal critical residues involved in activity. FEMS Microbiol. Lett. 325, 16–21. doi:10.1111/j.1574-6968.2011.02403.x
PubMed Abstract | CrossRef Full Text | Google Scholar
Mignouna, H. D., Abang, M. M., and Asiedu, R. (2008). in Genomics of yams, a common source of food and medicine in the tropics BT – genomics of tropical crop plants. Editors P. H. Moore, and R. Ming (New York: Springer), 549–570. doi:10.1007/978-0-387-71219-2_23
CrossRef Full Text | Google Scholar
Miller, J. C., and Tanksley, S. D. (1990). RFLP analysis of phylogenetic relationships and genetic variation in the genus Lycopersicon. TAG. Theor. Appl. Genet. Theor. Und Angewandte Genet. 80, 437–448. doi:10.1007/BF00226743
PubMed Abstract | CrossRef Full Text | Google Scholar
Montagnac, J. A., Davis, C. R., and Tanumihardjo, S. A. (2009). Nutritional value of cassava for use as a staple food and recent advances for improvement. Compr. Rev. Food Sci. Food Saf. 8, 181–194. doi:10.1111/j.1541-4337.2009.00077.x
PubMed Abstract | CrossRef Full Text | Google Scholar
Morris, K. J., Kamarulzaman, N. H., and Morris, K. I. (2019). Small-scale postharvest practices among plantain farmers and traders: a potential for reducing losses in rivers state, Nigeria. Sci. Afr. 4, e00086. doi:10.1016/j.sciaf.2019.e00086
CrossRef Full Text | Google Scholar
Muchira, N., Ngugi, K., Wamalwa, L. N., Avosa, M., Chepkorir, W., Manyasa, E., et al. (2021). Genotypic variation in cultivated and wild sorghum genotypes in response to Striga hermonthica infestation. Front. Plant Sci. 12, 671984. doi:10.3389/fpls.2021.671984
PubMed Abstract | CrossRef Full Text | Google Scholar
Mugabe, J. (2002). Biotechnology in sub-saharan Africa: towards a policy research agenda. Nairobi Kenya: African Technology Policy Studies Network, 1–28.
Google Scholar
Muhammad, R. H., Bamisheyi, E., and Olayemi, F. F. (2011). The effect of stage of ripening on the shelf life of tomatoes (Lycopersicon esculentum) stored in the evaporative cooling system (E.C.S). J. Dairy. Foods Home Sci. 30, 299–301.
Google Scholar
Mwanga, R. O. M., and Ssemakula, G. (2011). Orange-fleshed sweetpotatoes for food, health and wealth in Uganda. Int. J. Agric. Sustain. 9, 42–49. doi:10.3763/ijas.2010.0546
CrossRef Full Text | Google Scholar
Naik, P. L., Kotecha, M., Nathani, S., and Rathore, B. S. (2022). Rice-A review of nutritional and medicinal aspect mentioned in Ayurveda. South Asian Res. J. Pharm. Sci. 4, 1–5. doi:10.36346/sarjps.2022.v04i01.001
CrossRef Full Text | Google Scholar
Narayanan, N., Beyene, G., Chauhan, R. D., Gaitán-Solís, E., Gehan, J., Butts, P., et al. (2019). Biofortification of field-grown cassava by engineering expression of an iron transporter and ferritin. Nat. Biotechnol. 37, 144–151. doi:10.1038/s41587-018-0002-1
PubMed Abstract | CrossRef Full Text | Google Scholar
National Academies of Sciences, Engineering, and Medicine (2016). Genetically engineered crops: experiences and prospects. Washington, DC: The National Academies Press. doi:10.17226/23395
CrossRef Full Text | Google Scholar
Ndjeunga, J., and Nelson, C. H. (1999). Prospects for a pearl millet and sorghum food processing industry in West Africa semi-arid tropics. In: Towards sustainable sorghum production, utilization, and commercialization in West and central Africa proceedings of a technical workshop of the west and central Africa sorghum research network.
Google Scholar
Ndjiondjop, M. N., Semagn, K., Sie, M., Cissoko, M., Fatondji, B., and Jones, M. (2008). Molecular profiling of interspecific lowland rice populations derived from IR64 (Oryza sativa) and Tog5681 (Oryza glaberrima). Afr. J. Biotechnol. 7, 4219–4229.
Google Scholar
Ndjiondjop, M. N., Wambugu, P., Sangare, J. R., Dro, T., Kpeki, B., and Gnikoua, K. (2018). in Oryza glaberrima steud. BT – the wild Oryza genomes. Editors T. K. Mondal, and R. J. Henry (Springer International Publishing), 105–126. doi:10.1007/978-3-319-71997-9_9
CrossRef Full Text | Google Scholar
Neela, S., and Fanta, S. W. (2019). Review on nutritional composition of orange-fleshed sweet potato and its role in management of vitamin A deficiency. Food Sci. Nutr. 7, 1920–1945. doi:10.1002/fsn3.1063
PubMed Abstract | CrossRef Full Text | Google Scholar
Neelakandan, A. K., Subedi, B., Traore, S. M., Binagwa, P., Wright, D. A., and He, G. (2022). Base editing in peanut using CRISPR/nCas9. Front. Genome 4, 901444. doi:10.3389/fgeed.2022.901444
PubMed Abstract | CrossRef Full Text | Google Scholar
Nierman, W. C., Yu, J., Fedorova-Abrams, N. D., Losada, L., Cleveland, T. E., Bhatnagar, D., et al. (2015). Genome sequence of Aspergillus flavus NRRL 3357, a strain that causes aflatoxin contamination of food and feed. Genome announc. 3, 001688–e215. doi:10.1128/genomeA.00168-15
CrossRef Full Text | Google Scholar
Ning, J., He, W., Wu, L., Chang, L., Hu, M., Fu, Y., et al. (2023). The MYB transcription factor Seed Shattering 11 controls seed shattering by repressing lignin synthesis in African rice. Plant Biotechnol. J. 21, 931–942. doi:10.1111/pbi.14004
PubMed Abstract | CrossRef Full Text | Google Scholar
Njuguna, E., Coussens, G., Aesaert, S., Neyt, P., Anami, S., and Lijsebettens, M. V. (2017). Modulation of energy homeostasis in maize and Arabidopsis to develop lines tolerant to drought, genotoxic and oxidative stresses. Afr. Focus 30, 66–76. doi:10.21825/af.v30i2.8080
CrossRef Full Text | Google Scholar
Nkhata, S. G., Chilungo, S., Memba, A., and Mponela, P. (2020). Biofortification of maize and sweetpotatoes with provitamin A carotenoids and implication on eradicating vitamin A deficiency in developing countries. J. Agric. Food Res. 2, 100068. doi:10.1016/j.jafr.2020.100068
CrossRef Full Text | Google Scholar
Nyaboga, E., Tripathi, J. N., Manoharan, R., and Tripathi, L. (2014). Agrobacterium-mediated genetic transformation of yam (Dioscorea rotundata): an important tool for functional study of genes and crop improvement. Front. Plant Sci. 5 (463), 1–14. doi:10.3389/fpls.2014.00463
PubMed Abstract | CrossRef Full Text | Google Scholar
Odell, J. T., Nagy, F., and Chua, N. H. (1985). Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 313, 810–812. doi:10.1038/313810a0
PubMed Abstract | CrossRef Full Text | Google Scholar
Odipio, J., Alicai, T., Ingelbrecht, I., Nusinow, D. A., Bart, R., and Taylor, N. J. (2017). Efficient CRISPR/Cas9 genome editing of phytoene desaturase in cassava. Front. Plant Sci. 8, 1780. doi:10.3389/fpls.2017.01780
PubMed Abstract | CrossRef Full Text | Google Scholar
Ogbe, F. O., Dixon, A. G. O., Hughes, J. d.’A., Alabi, O. J., and Okechukwu, R. (2006). Status of cassava begomoviruses and their new natural hosts in Nigeria. Plant Dis. 90, 548–553. doi:10.1094/PD-90-0548
PubMed Abstract | CrossRef Full Text | Google Scholar
Ohkura, M., Cotty, P. J., and Orbach, M. J. (2018). Comparative genomics of Aspergillus flavus S and L morphotypes yield insights into niche adaptation. G3 (Bethesda) 8, 3915–3930. doi:10.1534/g3.118.200553
PubMed Abstract | CrossRef Full Text | Google Scholar
Okogbenin, E., Egesi, C. N., Olasanmi, B., Ogundapo, O., Kahya, S., Hurtado, P., et al. (2012). Molecular marker analysis and validation of resistance to cassava mosaic disease in elite cassava genotypes in Nigeria. N. Crop Sci. 52, 2576–2586. doi:10.2135/cropsci2011.11.0586
CrossRef Full Text | Google Scholar
Okogbenin, E., and Fregene, M. (2002). Genetic analysis and QTL mapping of early root bulking in an F1 population of non-inbred parents in cassava (Manihot esculenta Crantz). Theor. Appl. Genet. 106, 58–66. doi:10.1007/s00122-002-1068-0
PubMed Abstract | CrossRef Full Text | Google Scholar
Okogbenin, E., and Fregene, M. (2003). Genetic mapping of QTLs affecting productivity and plant architecture in a full-sib cross from non-inbred parents in Cassava (Manihot esculenta Crantz). Theor. Appl. Genet. 107, 1452–1462. doi:10.1007/s00122-003-1383-0
PubMed Abstract | CrossRef Full Text | Google Scholar
Okogbenin, E., Porto, M. C. M., Egesi, C., Mba, C., Ospinosa, E., Santos, L. G., et al. (2007). Marker-assisted introgression of resistance to cassava mosaic disease into Latin American germplasm for the genetic improvement of cassava in africa. Crop Sci. 47, 1895–1904. doi:10.2135/cropsci2006.10.0688
CrossRef Full Text | Google Scholar
Omeje, C. (2019). Media coverage and framing of genetically modified crops: a case study of science journalism in Nigeria. Stellenbosch, South Africa: Stellenbosch University. Masters Thesis.
Google Scholar
Onokpise, O. U., Wutoh, J. G., Ndzana, X., Tambong, J. T., Meboka, M. M., Sama, A. E., et al. (1999). “Evaluation of Macao cocoyam germplasm in Cameroon,” in Perspectives on new crops and new uses. Editor J. Janick (Alexandria, VA: ASHS Press), 394–396.
Google Scholar
Otekunrin, O. A., and Sawicka, B. (2019). Cassava, a 21st century staple crop: how can Nigeria harness its enormous trade potentials? Acta Sci. Agric. 3, 194–202. doi:10.31080/asag.2019.03.0586
CrossRef Full Text | Google Scholar
Oxfam (2019). Available in: gender inequalities and food insecurity. Oxfam, United Kingdom: Oxfam International.
Google Scholar
Oyekunle, M., Adamu, R. S., Ndou, E., Beyene, Y., Abdulmalik, M. M., and Oikeh, S. O. (2023). Efficacy of drought-tolerant and insect-protected transgenic TELA® maize traits in Nigeria. Transgenic Res. 32, 169–178. doi:10.1007/s11248-023-00345-x
PubMed Abstract | CrossRef Full Text | Google Scholar
Payne, G. A., Nierman, W. C., Wortman, J. R., Pritchard, B. L., Brown, D., Dean, R. A., et al. (2006). Whole genome comparison of Aspergillus flavus and A. oryzae. Med. Mycol. 44, S9-S11–S11. doi:10.1080/13693780600835716
PubMed Abstract | CrossRef Full Text | Google Scholar
Payne, G. A., Yu, J., Nierman, W. C., Machida, M., Bhatnagar, D., Cleveland, T. E., et al. (2008). “A first glance into the genome sequence of Aspergillus flavus,” in The aspergilli: genomics, medical aspects, biotechnology, and research methods. Editors S. A. Osmani, and G. H. Goldman (Boca Raton, FL, USA: CRC Press), 15–23.
Google Scholar
Pellegrino, E., Bedini, S., Nuti, M., and Ercoli, L. (2018). Impact of genetically engineered maize on agronomic, environmental and toxicological traits: a meta-analysis of 21 years of field data. Sci. Rep. 8, 3113. doi:10.1038/s41598-018-21284-2
PubMed Abstract | CrossRef Full Text | Google Scholar
Pérez-Vich, B., Velasco, L., Rich, P. J., and Ejeta, G. (2013). in Marker-assisted and physiology-based breeding for resistance to root parasitic Orobanchaceae BT – parasitic Orobanchaceae: parasitic mechanisms and control strategies. Editors D. M. Joel, J. Gressel, and L. J. Musselman (Springer Berlin Heidelberg), 369–391. doi:10.1007/978-3-642-38146-1_21
CrossRef Full Text | Google Scholar
Poutanen, K. S., Kårlund, A. O., Gómez-Gallego, C., Johansson, D. P., Scheers, N. M., Marklinder, I. M., et al. (2022). Grains – a major source of sustainable protein for health. Nutr. Rev. 80, 1648–1663. doi:10.1093/nutrit/nuab084
PubMed Abstract | CrossRef Full Text | Google Scholar
Prasad, K., Yogendra, K., Sanivarapu, H., Kanniah, R., Cary, J. W., Sharma, K. K., et al. (2023). Multiplexed host-induced gene silencing of Aspergillus flavus. toxins Toxins (Basel). 15, 319. doi:10.3390/toxins15050319
PubMed Abstract | CrossRef Full Text | Google Scholar
Ragaee, S., Abdel-Aal, E.-S. M., and Noaman, M. (2006). Antioxidant activity and nutrient composition of selected cereals for food use. Food Chem. 98, 32–38. doi:10.1016/j.foodchem.2005.04.039
CrossRef Full Text | Google Scholar
Ramu, P., Srivastava, R. K., Sanyal, A., Fengler, K., Cao, J., Zhang, Y., et al. (2023). Improved pearl millet genomes representing the global heterotic pool offer a framework for molecular breeding applications. Commun. Biol. 6 (1), 902. doi:10.1038/s42003-023-05258-3
PubMed Abstract | CrossRef Full Text | Google Scholar
Rasmussen, A., Mason, M. G., De Cuyper, C., Brewer, P. B., Herold, S., Agusti, J., et al. (2012). Strigolactones suppress adventitious rooting in Arabidopsis and pea. Plant Physiol. 158, 1976–1987. doi:10.1104/pp.111.187104
PubMed Abstract | CrossRef Full Text | Google Scholar
Redinbaugh, M. G., and Stewart, L. R. (2018). Maize Lethal Necrosis: an emerging, synergistic viral disease. Annu. Rev. Virology 5, 301–322. doi:10.1146/annurev-virology-092917-043413
PubMed Abstract | CrossRef Full Text | Google Scholar
Rich, P. J., and Ejeta, G. (2007). “Biology of host-parasite interactions in striga species,” in Integrating new technologies for Striga control (World Scientific), 19–32. doi:10.1142/9789812771506_0002
CrossRef Full Text | Google Scholar
Rodríguez, G. R., Pratta, G. R., Zorzoli, R., and Picardi, L. A. (2006). Recombinant lines obtained from an interspecific cross between Lycopersicon Species selected by fruit weight and fruit shelf life. J. Am. Soc. Hortic. Sci. Jashs 131, 651–656. doi:10.21273/JASHS.131.5.651
CrossRef Full Text | Google Scholar
Rudi, N., Norton, G. W., Alwang, J., and Asumugha, G. (2010). Economic impact analysis of marker-assisted breeding for resistance to pests and postharvest deterioration in cassava. Afr. J. Agric. Resour. Econ. 4, 110–122. doi:10.22004/ag.econ.93862
CrossRef Full Text | Google Scholar
Samodien, E., Jewell, J. F., Loedolff, B., Oberlander, K., George, G. M., Zeeman, S. C., et al. (2018). Repression of SEX4 and LIKE SEX FOUR2 orthologs in potato increases tuber starch bound phosphate with concomitant alterations in starch physical properties. Front. Plant Sci. 9, 1044. doi:10.3389/fpls.2018.01044
PubMed Abstract | CrossRef Full Text | Google Scholar
Santelia, D., and Zeeman, S. C. (2011). Progress in Arabidopsis starch research and potential biotechnological applications. Curr. Opin. Biotechnol. 22, 271–280. doi:10.1016/j.copbio.2010.11.014
PubMed Abstract | CrossRef Full Text | Google Scholar
Sarla, N., and Swamy, B. P. M. (2005). Oryza glaberrima: a source for the improvement of Oryza sativa. Curr. Sci. 89, 955–963.
Google Scholar
Satish, L., Ceasar, S. A., and Ramesh, M. (2017). Improved Agrobacterium mediated transformation and direct plant regeneration in four cultivars of fnger millet (Eleusine coracana (L.) Gaertn.). Plant Cell. Tissue Organ Cult. 131, 547–565. doi:10.1007/s11240-017-1305-5
CrossRef Full Text | Google Scholar
Sayre, R., Beeching, J. R., Cahoon, E. B., Egesi, C., Fauquet, C., Fellman, J., et al. (2011). The BioCassava plus program: biofortification of cassava for sub-saharan africa. Annu. Rev. Plant Biol. 62, 251–272. doi:10.1146/annurev-arplant-042110-103751
PubMed Abstract | CrossRef Full Text | Google Scholar
Schussler, J. R., and Westgate, M. E. (1995). Assimilate flux determines kernel set at low water potential in maize. Crop Sci. 35, 1074–1080. doi:10.2135/cropsci1995.0011183X003500040026x
CrossRef Full Text | Google Scholar
Schwartzbeck, J. L., Jung, S., Abbott, A. G., Mosley, E., Lewis, S., Pries, G. L., et al. (2001). Endoplasmic oleoyl-PC desaturase references the second double bond. Phytochemistry 57, 643–652. doi:10.1016/s0031-9422(01)00081-4
PubMed Abstract | CrossRef Full Text | Google Scholar
Senay, S. D., Pardey, P. G., Chai, Y., Doughty, L., and Day, R. (2022). Fall armyworm from a maize multi-peril pest risk perspective. Front. Insect Sci. 2, 971396. doi:10.3389/finsc.2022.971396
PubMed Abstract | CrossRef Full Text | Google Scholar
Shah, D. M., Horsch, R. B., Klee, H. J., Kishore, G. M., Winter, J. A., Tumer, N. E., et al. (1986). Engineering herbicide tolerance in transgenic plants. Sci. (New York, N.Y.) 233, 478–481. doi:10.1126/science.233.4762.478
PubMed Abstract | CrossRef Full Text | Google Scholar
Sharma, K. K., Pothana, A., Prasad, K., Shah, D., Kaur, J., Bhatnagar, D., et al. (2018). Peanuts that keep aflatoxin at bay: a threshold that matters. Plant Biotechnol. J. 16 (5), 1024–1033. doi:10.1111/pbi.12846
PubMed Abstract | CrossRef Full Text | Google Scholar
Sharma, M., Yadav, D. N., Singh, A. K., and Tomar, S. K. (2015). Rheological and functional properties of heat moisture treated pearl millet starch. J. Food Sci. Technol. 52, 6502–6510. doi:10.1007/s13197-015-1735-1
PubMed Abstract | CrossRef Full Text | Google Scholar
Sisay, B., Tefera, T., Wakgari, M., Ayalew, G., and Mendesil, E. (2019). The efficacy of selected synthetic insecticides and botanicals against fall armyworm, Spodoptera frugiperda, in Maize. Insects 10, 45. doi:10.3390/insects10020045
PubMed Abstract | CrossRef Full Text | Google Scholar
Smith, A. M., and Zeeman, S. C. (2020). Starch: a flexible, adaptable carbon store coupled to plant growth. Annu. Rev. Plant Biol. 71, 217–245. doi:10.1146/annurev-arplant-050718-100241
PubMed Abstract | CrossRef Full Text | Google Scholar
Sonnewald, U., Fernie, A. R., Gruissem, W., Schläpfer, P., Anjanappa, R. B., Chang, S. H., et al. (2020). The Cassava Source–Sink project: opportunities and challenges for crop improvement by metabolic engineering. Plant J. 103, 1655–1665. doi:10.1111/tpj.14865
PubMed Abstract | CrossRef Full Text | Google Scholar
Sood, P., Singh, R. K., and Prasad, M. (2019). Millets genetic engineering: the progress made and prospects for the future. Plant Cell. Tiss. Organ Cult. 137, 421–439. doi:10.1007/s11240-019-01587-6
CrossRef Full Text | Google Scholar
Spillane, C., and Gepts, P. (2001). Evolutionary and genetic perspectives on the dynamics of crop genepools. International Plant Genetic Resources Institute (IPGRI) Food and Agriculture Organization of the United Nations (FAO) CABI (25–70). doi:10.1079/9780851994116.0025
CrossRef Full Text | Google Scholar
Statista, (2024). Available at: Nigeria: people in extreme poverty 2016-2025 | Statista
Google Scholar
Stevens, G. A., Beal, T., Mbuya, M. N. N., Luo, H., and Neufeld, L. M.Global Micronutrient Deficiencies Research Group (2022). Micronutrient deficiencies among preschool-aged children and women of reproductive age worldwide: a pooled analysis of individual-level data from population-representative surveys. Lancet. Glob. Health 10, e1590–e1599. doi:10.1016/S2214-109X(22)00367-9
PubMed Abstract | CrossRef Full Text | Google Scholar
Stevens, P. F. (2012). Angiosperm phylogeny website. Version 13. St Louis, MO: Missouri Botanical Garden, University of Missouri.
Google Scholar
Swaminaidu, N., Ghosh, S., and Mallikarjuna, K. (2015). Millets: the miracle grain. Int. J. Pharma. Bio. Sci. 6, 440–446.
Google Scholar
Syombua, E. D., Zhang, Z., Tripathi, J. N., Ntui, V. O., Kang, M., George, O. O., et al. (2021). A CRISPR/Cas9-based genome-editing system for yam (Dioscorea spp.). Plant Biotechnol. J. 19, 645–647. doi:10.1111/pbi.13515
PubMed Abstract | CrossRef Full Text | Google Scholar
Tambong, J. T., and Meboka, M. (1994). Cocoyam (Xanthosoma sagittifolium (L.)) hybridization studies: pollen viability and seed germination. Acta Hortic. 380, 448–459. doi:10.17660/ActaHortic.1994.380.69
CrossRef Full Text | Google Scholar
Taylor, N., Gaitán-Solís, E., Moll, T., Trauterman, B., Jones, T., Pranjal, A., et al. (2012). A high-throughput platform for the production and analysis of transgenic cassava (Manihot esculenta) plants. Trop. Plant Biol. 5 (2012), 127–139. doi:10.1007/s12042-012-9099-4
CrossRef Full Text | Google Scholar
Taylor, N., Paul, C., Raemakers, K., Siritunga, D., and Zhang, P. (2004). Development and application of transgenic technologies in cassava. Plant Mol. Biol. 56, 671–688. doi:10.1007/s11103-004-4872-x
PubMed Abstract | CrossRef Full Text | Google Scholar
Taylor, N. J., Halsey, M., Gaitán-Solís, E., Anderson, P., Gichuki, S., Miano, D., et al. (2012). The VIRCA Project: virus resistant cassava for Africa. GM Crops Food 3 (2), 93–103. doi:10.4161/gmcr.19144
PubMed Abstract | CrossRef Full Text | Google Scholar
Teklu, D. H., Shimelis, H., and Abady, S. (2022). Genetic improvement in Sesame (Sesamum indicum L.): progress and outlook: a review. Agronomy 12, 2144. doi:10.3390/agronomy12092144
CrossRef Full Text | Google Scholar
The Conversation (2022). South Africa should rethink regulations on genetically modified plants (theconversation.com).
Google Scholar
The Conversation (2023). Nigeria is Africa’s leading rice producer, but still needs more – reusing wastewater for irrigation would boost farming (theconversation.com).
Google Scholar
The Royal Society (1999). Available at: review of data on possible toxicity of GM potatoes. United Kingdom: Royal Society.
Google Scholar
Thottappilly, G., and Loebenstein, G. (2009). “Concluding remarks,” in The sweetpotato. Editors G. Loebenstein, and G. Thottappilly (Dordrecht: Springer). doi:10.1007/978-1-4020-9475-0_23
CrossRef Full Text | Google Scholar
Tiwari, S., Kumar, S. V., and Gontia, I. (2011). Minireview: biotechnological approaches for sesame (Sesamum indicum L.) and Niger (Guizotia abyssinica L.f. Cass.). Asia-Pacific J. Mol. Biol. Biotechnol. 19, 2–9.
Google Scholar
Tripathi, J. N., Ntui, V. O., Ron, M., Muiruri, S. K., Britt, A., and Tripathi, L. (2019). CRISPR/Cas9 editing of endogenous banana streak virus in the B genome of Musa spp. overcomes a major challenge in banana breeding. Commun. Biol. 2, 46. doi:10.1038/s42003-019-0288-7
PubMed Abstract | CrossRef Full Text | Google Scholar
Tukura, D., and Ashindo, E. (2019). Determinant of technical efficiency of Sesame production in Kurmi local government area of Taraba State, Nigeria. IOSR J. Agric. Veterinary Sci. 12, 43–51. doi:10.9790/2380-1205014351
CrossRef Full Text | Google Scholar
Tumuhimbise, A. G., Orishaba, J., Atukwase, A., and Namutebi, A. (2013). Effect of salt on the sensory and keeping quality of orange fleshed sweet potato crisps. Food Nutr. Sci. J. 4, 454–460. doi:10.4236/fns.2013.44058
CrossRef Full Text | Google Scholar
Ubalua, A. O., Ewa, F., and Okeagu, O. D. (2016). Potentials and challenges of sustainable Taro (Colocasia esculenta) production in Nigeria. J. App. Biol. Biotech. 4, 53–59. doi:10.7324/JABB.2016.40110
CrossRef Full Text | Google Scholar
Ugonna, C., and Onwualu, P. A. (2015). Tomato value chain in Nigeria: issues, challenges and strategies. J. Sci. Res. Rep. 7, 501–515. doi:10.9734/JSRR/2015/16921
CrossRef Full Text | Google Scholar
Umar, U. A., Muntaqa, A. H., Muhammad, M. B., and Jantar, H. J. (2014). Review of Sesame seed production and export in Nigeria (2003 to 2012). Pac. J. Sci. Technol. 15, 200–203.
Google Scholar
United Nations Children’s Fund [UNICEF] (2023). “Undernourished and overlooked: a global nutritional crisis in adolescent girls and women,” in UNICEF child nutrition report series, 2022. New York: UNICEF.
Google Scholar
Upadhyaya, H. D., Gowda, C. L., Pundir, R. P., Reddy, V. G., and Singh, S. (2006). Development of core subset of finger millet germplasm using geographical origin and data on 14 quantitative traits. Genet. Resour. Crop Evol. 53 (4), 679–685. doi:10.1007/s10722-004-3228-3
CrossRef Full Text | Google Scholar
Varshney, R. K., Shi, C., Thudi, M., Mariac, C., Wallace, J., Qi, P., et al. (2017). Pearl millet genome sequence provides a resource to improve agronomic traits in arid environ-ments. Nat. Biotechnol. 35, 969–976. doi:10.1038/nbt.3943
PubMed Abstract | CrossRef Full Text | Google Scholar
Veley, K. M., Okwuonu, I., Jensen, G., Yoder, M., Taylor, N. J., Meyers, B. C., et al. (2021). Gene tagging via CRISPR-mediated homology-directed repair in cassava. G3 Genes.|Genomes|Genetics 11 (4), jkab028–9. doi:10.1093/g3journal/jkab028
CrossRef Full Text | Google Scholar
Wal, J. M. (2015). “Assessing and managing allergenicity of genetically modified (GM) foods,” in Handbook of food allergen detection and control. Editor S. Flanagan (Cambridge, UK: Woodhead Publishing), 161–178.
CrossRef Full Text | Google Scholar
Wambugu, P. W., Furtado, A., Waters, D. L. E., Nyamongo, D. O., and Henry, R. J. (2013). Conservation and utilization of African Oryza genetic resources. Rice (New York, N.Y.) 6, 29. doi:10.1186/1939-8433-6-29
PubMed Abstract | CrossRef Full Text | Google Scholar
Wambugu, P. W., Ndjiondjop, M. N., and Henry, R. (2019). Advances in molecular genetics and genomics of African rice (Oryza glaberrima Steud). Plants Basel, Switz. 8, 376. doi:10.3390/plants8100376
PubMed Abstract | CrossRef Full Text | Google Scholar
Wang, D., Samsulrizal, N. H., Yan, C., Allcock, N. S., Craigon, J., Blanco-Ulate, B., et al. (2019). Characterization of CRISPR mutants targeting genes modulating pectin degradation in ripening tomato. Plant Physiol. 179, 544–557. doi:10.1104/pp.18.01187
PubMed Abstract | CrossRef Full Text | Google Scholar
Wang, H., Wu, Y., Zhang, Y., Yang, J., Fan, W., Zhang, H., et al. (2019). CRISPR/Cas9-Based mutagenesis of starch biosynthetic genes in sweet potato (Ipomoea batatas) for the improvement of starch quality. Int. J. Mol. Sci. 20 (19), 4702. doi:10.3390/ijms20194702
PubMed Abstract | CrossRef Full Text | Google Scholar
Wang, Q., Liu, Y., Xie, Y., and You, M. (2006). Cryotherapy of potato shoot tips for efficient elimination of potato leafroll virus (PLRV) and potato virus Y (PVY). Potato Res. 49, 119–129. doi:10.1007/s11540-006-9011-4
CrossRef Full Text | Google Scholar
Wang, Q. C., and Valkonen, J. P. T. (2008). Elimination of two viruses which interact synergistically from sweetpotato by shoot tip culture and cryotherapy. J. Virological Methods 154, 135–145. doi:10.1016/j.jviromet.2008.08.006
PubMed Abstract | CrossRef Full Text | Google Scholar
Wang, W., Hostettler, C. E., Damberger, F. F., Kossmann, J., Lloyd, J. R., and Zeeman, S. C. (2018). Modification of cassava root starch phosphorylation enhances starch functional properties. Front. Plant Sci. 9, 1562. doi:10.3389/fpls.2018.01562
PubMed Abstract | CrossRef Full Text | Google Scholar
Wei, X., Liu, K., Zhang, Y., Feng, Q., Wang, L., Zhao, Y., et al. (2015). Genetic discovery for oil production and quality in sesame. Nat. Commun. 6, 8609. doi:10.1038/ncomms9609
PubMed Abstract | CrossRef Full Text | Google Scholar
Wei, X., Zhu, X., Yu, J., Wang, L., Zhang, Y., Li, D., et al. (2016). Identification of sesame genomic variations from genome comparison of landrace and variety. Front. Plant Sci. 7, 1169. doi:10.3389/fpls.2016.01169
PubMed Abstract | CrossRef Full Text | Google Scholar
Wilson, D. M., and Stansell, J. R. (1983). Effect of irrigation regimes on aflatoxin contamination of peanut pods. Peanut Sci. 10, 54–56. doi:10.3146/i0095-3679-10-2-2
CrossRef Full Text | Google Scholar
World Health Organization (2008). The global burden of disease: 2004 update. Geneva: WHO Press.
Google Scholar
Wu, S., Lau, K. H., Cao, Q., Hamilton, J. P., Sun, H., Zhou, C., et al. (2018). Genome sequences of two diploid wild relatives of cultivated sweet potato reveal targets for genetic improvement. Nat. Commun. 9, 4580. doi:10.1038/s41467-018-06983-8
PubMed Abstract | CrossRef Full Text | Google Scholar
Yadav, M., Chaudhary, D., Sainger, M., and Jaiwal, P. K. (2010). Agrobacterium tumefaciens-mediated genetic transformation of sesame (Sesamum indicum L.). Plant Cell., Tissue Organ Cult. (PCTOC) 103, 377–386. doi:10.1007/s11240-010-9791-8
CrossRef Full Text | Google Scholar
Yang, B. (2005). Mendel in the kitchen: a scientist’s view of genetically modified foods. Discov. Med. 5, 324–330.
PubMed Abstract | Google Scholar
Yang, L., Huang, W., Xiong, F., Xian, Z., Su, D., Ren, M., et al. (2017). Silencing of SlPL, which encodes a pectate lyase in tomato, confers enhanced fruit firmness, prolonged shelf-life and reduced susceptibility to grey mould. Plant Biotechnol. J. 15, 1544–1555. doi:10.1111/pbi.12737
PubMed Abstract | CrossRef Full Text | Google Scholar
Yu, B., Jiang, H., Pandey, M. K., Huang, L., Huai, D., Zhou, X., et al. (2020). Identification of two novel peanut genotypes resistant to aflatoxin production and their SNP markers associated with resistance. Toxins 12 (3), 156. doi:10.3390/toxins12030156
PubMed Abstract | CrossRef Full Text | Google Scholar
Yuan, M., Zhu, J., Gong, L., He, L., Lee, C., Han, S., et al. (2019). Mutagenesis of FAD2 genes in peanut with CRISPR/Cas9 based gene editing. BMC Biotechnol. 19 (24), 24. doi:10.1186/s12896-019-0516-8
PubMed Abstract | CrossRef Full Text | Google Scholar
Zainuddin, I. M., Fathoni, A., Sudarmonowati, E., Beeching, J. R., Gruissem, W., and Vanderschuren, H. (2018). Cassava post-harvest physiological deterioration: from triggers to symptoms. Postharvest Biol. Technol. 142, 115–123. doi:10.1016/j.postharvbio.2017.09.004
CrossRef Full Text | Google Scholar
Zaknayiba, D. B., and Tanko, L. (2013). Costs and returns analysis of yam production among small scale farmers in Karu local government area, Nasarawa State, Nigeria. PAT 9, 73–80.
Google Scholar
Zeeman, S. C., Kossmann, J., and Smith, A. M. (2010). Starch: its metabolism, evolution, and biotechnological modification in plants. Annu. Rev. Plant Biol. 61, 209–234. doi:10.1146/annurev-arplant-042809-112301
PubMed Abstract | CrossRef Full Text | Google Scholar
Zhang, L., Zhao, L., Bian, X., Guo, K., Zhou, L., and Wei, C. (2018). Characterization and comparative study of starches from seven purple sweet potatoes. Food Hydrocoll. 80, 168–176. doi:10.1016/j.foodhyd.2018.02.006
CrossRef Full Text | Google Scholar
Zhang, L., Zhu, M., Ren, L., Li, A., Chen, G., and Hu, Z. (2018). The SlFSR gene controls fruit shelf-life in tomato. J. Exp. Bot. 69, 2897–2909. doi:10.1093/jxb/ery116
PubMed Abstract | CrossRef Full Text | Google Scholar
Zhang, L. P., Khan, A., Niño-Liu, D., and Foolad, M. R. (2002). A molecular linkage map of tomato displaying chromosomal locations of resistance gene analogs based on a Lycopersicon esculentum x Lycopersicon hirsutum cross. Genome 45, 133–146. doi:10.1139/g01-124
PubMed Abstract | CrossRef Full Text | Google Scholar
Zhang, P., Vanderschuren, H., Fütterer, J., and Gruissem, W. (2005). Resistance to cassava mosaic disease in transgenic cassava expressing antisense RNAs targeting virus replication genes. Plant Biotechnol. J. 3, 385–397. doi:10.1111/j.1467-7652.2005.00132.x
PubMed Abstract | CrossRef Full Text | Google Scholar
Zhao, X., Jayarathna, S., Turesson, H., Fält, A. S., Nestor, G., González, M. N., et al. (2021). Amylose starch with no detectable branching developed through DNA-free CRISPR-Cas9 mediated mutagenesis of two starch branching enzymes in potato. Sci. Rep. 11, 4311. doi:10.1038/s41598-021-83462-z
PubMed Abstract | CrossRef Full Text | Google Scholar
Zhong, Y., Blennow, A., Kofoed-Enevoldsen, O., Jiang, D., and Hebelstrup, K. H. (2019). Protein Targeting to Starch 1 is essential for starchy endosperm development in barley. J. Exp. Bot. 70, 485–496. doi:10.1093/jxb/ery398
PubMed Abstract | CrossRef Full Text | Google Scholar
Zhou, W., He, S., Naconsie, M., Ma, Q., Zeeman, S. C., Gruissem, W., et al. (2017). Alpha-glucan, water dikinase 1 affects starch metabolism and storage root growth in cassava (Manihot esculenta Crantz). Sci. Rep. 7, 9863. doi:10.1038/s41598-017-10594-6
PubMed Abstract | CrossRef Full Text | Google Scholar
Zhuang, W., Chen, H., Yang, M., Wang, J., Pandey, M. K., Zhang, C., et al. (2019). The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 51 (5), 865–876. doi:10.1038/s41588-019-0402-2
PubMed Abstract | CrossRef Full Text | Google Scholar
Zsögön, A., Čermák, T., Naves, E. R., Notini, M. M., Edel, K. H., Weinl, S., et al. (2018). De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 36, 1211–1216. doi:10.1038/nbt.4272
CrossRef Full Text | Google Scholar
Glossary
Source link : https://www.frontiersin.org/journals/genome-editing/articles/10.3389/fgeed.2024.1398813/full
Author :
Publish date : 2024-06-30 16:31:01
Copyright for syndicated content belongs to the linked Source.