Introduction
The Lesotho Highlands, primarily located in Lesotho and marginal parts of South Africa (and which includes a large portion of the World Heritage, transfrontier Maloti-Drakensberg Park), are some of the highest elevated land in Africa (maximum elevation = 3,482 m.a.s.l.) which maintain a particularly cooler climate against the backdrop of the broader, temperate southern African region (Norström et al., 2018; Hoogendoorn et al., 2020; Figure 1A). Although it is part of the Drakensberg mountain range and the Great Escarpment, which stretches from the Eastern Cape to Limpopo, the Lesotho Highlands are isolated by being ∼1,500 m higher due to Jurassic vulcanism which led to the extra basaltic cover giving the highland its pronounced elevation (see Eriksson, 1981; Smith et al., 1993). The Highlands act as the catchment and water tower for several large drainages that filter off in different directions across the southern African landscape. As such, the region holds a diverse array of freshwater environments from rivers and streams to ponds and wetlands (Norström et al., 2018; Figures 1B,C), but species richness is virtually unknown for most aquatic groups. Similar cooler, high-altitude regions in temperate landscapes have been shown to support high species richness due to acting as a melting pot of museums (i.e., refugia) for freshwater fauna that evolved under the Pleistocene glacial periods (and require cooler conditions to survive—i.e.,“cold-evolved” species), as well as sinks for opportunistic, temperate species that are expanding their ranges post the Last Glacial Maximum (Füreder et al., 2006; Rahbek et al., 2019; Hågvar et al., 2020; Brighenti et al., 2021; Clewing et al., 2022). As such, similar has been predicted for the Lesotho Highlands aquatic environments (Skelton, 2000) and the region could therefore be important for the conservation of freshwater biota (certainly elevated diversity and/or high endemism has been shown in several terrestrial groups, including land molluscs; Hamer and Slotow, 2009; Kopij, 2015; Perera et al., 2021), but it is equally possible that the generally reduced dispersal capabilities of aquatic fauna in conjunction with glacial cycles, if these were dramatic enough, could have caused several turnover events—where the current species assemblages are all the result of very recent colonisations, thus being relatively species poor.
FIGURE 1. (A) Lesotho Highlands covered in winter snow from space (NASA Earth Observatory images by Lauren Dauphin, using MODIS data from LANCE/EOSDIS Rapid Response), (B,C) Typical freshwater streams in the Lesotho Highlands and (D) Burnupia attached to rocks in a Lesotho Highland stream.
In the case of freshwater species conservation in the Lesotho Highlands, knowing if there is indeed high relative species richness and cold-evolved species is paramount given growing anthropogenic activity (e.g., waste pollution and agriculture; see Pullanikkatil and Urama, 2011; Chatanga and Seleteng-Kose, 2021; Turpie et al., 2021) and global warming—this is particularly relevant for southern Africa and cold-evolved species, where climatic change is likely to increase substantially over the next century (Dallas and Rivers-Moore, 2014; Serdeczny et al., 2016; Archer et al., 2018; Bentley et al., 2018; Weber et al., 2018). However, conservation relies on accurately being able to quantify relative biodiversity richness in the Highlands as compared with the surroundings and, in this case, decipher cold-evolved species from not (i.e., through determining the evolutionary and spatial origins of species). Molecular phylogenetic techniques can provide a suitable way of establishing the above when dated with molecular clocks and can circumvent some of the pitfalls quantifying species richness through traditional morphology-based methods (e.g., which often have the inability to identify cryptic species). Sadly, there has been a neglect of modern molecular evolutionary aquatic research in the Lesotho Highlands and few studies exist that can even attempt to address these questions in freshwater biota (e.g., Daniels et al., 2003; Swartz, 2005; Swartz et al., 2009; Tolley et al., 2010; Phiri et al., 2016).
In the absence of suitable literature and having molecular evolutionary data for a multitude of species, reconstructing the evolutionary history of a model group may help provide some indications on the relative levels, origins and types of diversity in the Lesotho Highlands. A model taxon needs to be well distributed across southern Africa, have several species, lack a free-swimming larval stage to avoid dispersal biases associated with currents or flow and be a running-water species to avoid dispersal and temporal biases that may be caused by species that occur in transient or short-lived aquatic environments. In this regard, the freshwater limpet genus Burnupia meets all the criteria (Figure 1D): They are found in high-oxygenated, moving waters from the Western Cape in South Africa right up, through Lesotho, into Botswana, Zimbabwe, Mozambique, Eswatini and even further into the burbling littoral zones of the great lakes of eastern Africa (Brown, 1994; de Kock and Wolmarans, 2009, 2016, 2017; Albrecht and Clewing, 2019). Moreover, there are 22 morphospecies described from across Africa (with 15 occurring specifically within the southern African region; Connolly, 1939; Brown, 1994; Figure 2A) which lay a stationary egg mass from which fully formed juvenile limpets emerge (Davies-Coleman, 2001).
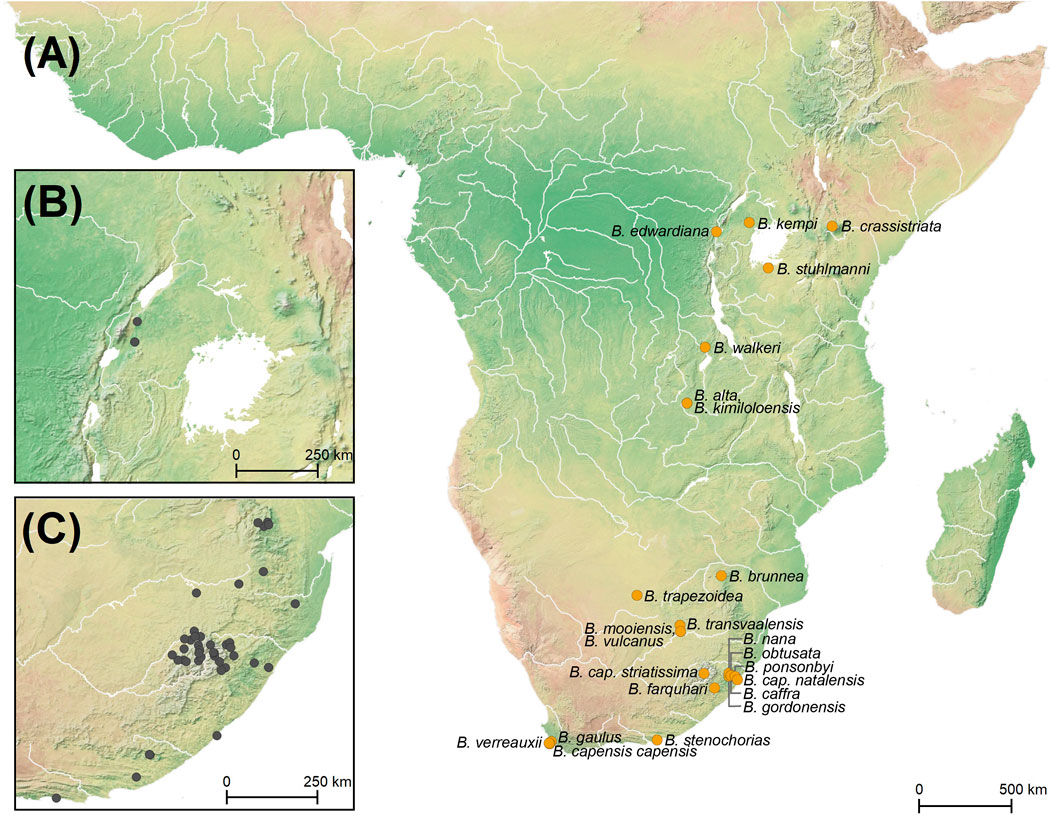
FIGURE 2. (A) Map of Africa with the type localities of 22 Burnupia species and three subspecies of Burnupia capensis marked in gold dots, with sub maps showing (B) the sampling localities of Burnupia incorporated into the current study from eastern Africa and (C) southern Africa (inclusive of the Lesotho Highlands) in grey dots.
In this study, we sampled the genus Burnupia across southern and eastern Africa to gain evolutionary insights into the relative freshwater biodiversity of the Lesotho Highlands using molecular phylo- and population-genetic approaches. Specifically, we aimed to; 1) determine if molecular species diversity is rich in the Highlands in comparison to the broader region, 2) assess the evolutionary history of the genus to determine their origin in/colonisation history of the Highlands and the degree of endemism occurring there, as well as, 3) establish if cold-evolved species exist for the genus in the Highlands. To address these aims, we first reconstruct the evolutionary history of the genus using multiple phylogenetic approaches and place a temporal perspective on divergence events using an established molecular clock rate. Secondly, we delimit phylospecies to determine the diversity in the Highlands and surroundings. Thirdly, we determine the temporal and spatial origin and/or colonisation history of Highland species by modelling ancestral area estimations and, finally, assess for cold-evolved species therein. As these outcomes and methodological approaches generate a lot of data useful for the Burnupia systematics, and several authors have encouraged a molecular review of the genus (Brown, 1994; Albrecht et al., 2004; de Kock and Wolmarans, 2016, 2017; Albrecht and Clewing, 2019), we also discuss the taxonomic implications of our work.
It is hoped that the outcomes of this research will provide us with a preliminary indication of when and where the Highland’s freshwater diversity originated and if it is an important area for conservation consideration in respect to relative diversity and/or cold-evolved species. Moreover, we hope the results will encourage and provide guidance and direction for freshwater biogeographic research in the Highlands going forward.
MethodsSampling Design, Laboratory Protocols and Sequence Alignment
Forty-five Burnupia specimens were hand-collected from hard substrate in shallow water streams and river pools and dams (e.g., Figures 1D, 2B,C). Protancylus pileolus was used as an outgroup to root downstream phylogenetic analyses (Table 1). Specimens were preserved in 80% ethanol, before being utilised for DNA extraction. Emphasis was placed on collecting Burnupia from as many locations across the Lesotho Highlands and surrounding regions as possible (Figures 2B,C; Table 1). Genomic DNA was extracted from the foot tissue of each specimen using a DNeasy Blood and Tissue kit (QIAGEN) or via the CTAB extraction method (Winnepenninckx, 1993). Amplification occurred for a 655 bp region of the mitochondrial DNA cytochrome c oxidase subunit 1 (COI) gene through PCR with the primers LCO1490 (Folmer et al., 1994) and COR722b (Wilke and Davis, 2000) and under the following cyclic conditions: 95°C—5 min; 30 cycles of 95°C—10 s, 40°C—30 s, 70°C—1 min; final elongation at 72°C—10 min. The success of PCR amplifications was confirmed by 1% agarose gel electrophoresis, before purification of gene fragments and bidirectional Sanger sequencing were carried out by LGC Ltd. (Berlin, Germany). Sequence ends were trimmed in Geneious 10.1.2 (Biomatters Ltd., 2017). To expand our dataset, all additional sequences from Burnupia specimens with corresponding COI data published on GenBank (www.ncbi.nlm.nih.gov/genbank) where incorporated (i.e., two sequences; Table 1). Alignments of the 48 gene fragments were performed using Geneious 10.1.2 and the Geneious alignment algorithm.
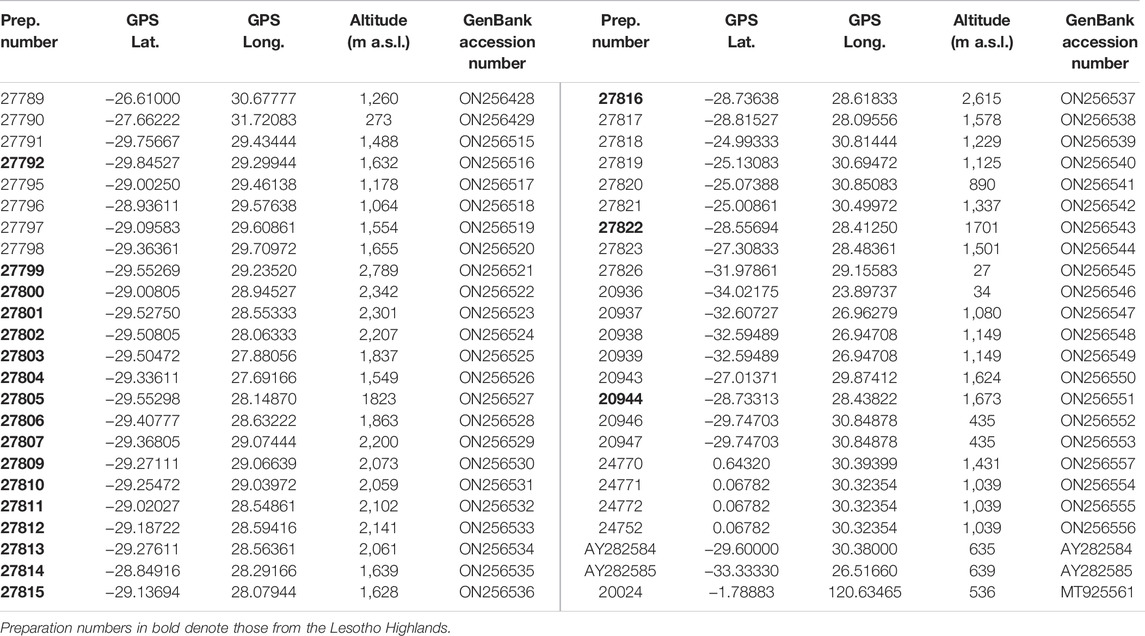
TABLE 1. Preparation numbers, GPS coordinates (Longitude and Latitude), altitude and GenBank accession numbers of all Burnupia specimens used in the current study and the Protancylus outgroup (Prep. number: 20024).
Phylogenetic Analyses and Molecular Dating
To reconstruct the evolutionary history of Burnupia, phylogenetic relationships were established following the methods of Sands et al. (2019a) with only minor differences. In summary, PAUP 4.0b (Swofford, 2002) and MrBayes 3.2.6 (Ronquist et al., 2012) were used to generate unweighted maximum parsimony (MP) and model-informed Bayesian inference (BI) based phylogenies respectively conducted through the CIPRES Science Gateway (Miller et al., 2010). Firstly, the MP analysis was constructed using the heuristic search function, with TBR branch swapping and 100 random taxon additions. Only the best tree was saved during each replicate and the robustness of nodes were assessed by 10,000 bootstrap (BS) replicates (where values ≥70% were considered supported; Felsenstein, 1985). Secondly, the BI analysis was performed to determine the posterior probabilities (PP) of associations. The HKY+I+Γ was selected as the best-fit model of sequence evolution (determined using jModelTest 2.1.10; Darriba et al., 2012) using the Akaike Information Criterion (AIC; Akaike, 1973) and two parallel Markov Chain Monte Carlo (MCMC) simulations used five chains for 100,000,000 generations, saving one tree in every 2,000 generations. The BI trees were summarised after the first 20% of trees were discarded as burn-in, as assessed by parameter convergence in Tracer 1.6 (Rambaut et al., 2014). Nodes with PP ≥ 0.95 where considered supported. Finally, as the BI and MP topologies were highly comparable and no conflict was found between supported nodes of these methods, BS values ≥70% from the MP phylogeny (phylogeny not shown) where transferred onto the BI phylogeny.
To place a temporal perspective on divergence events, to observe when the Lesotho Highlands were colonised and if these overlap with major climatic or geological changes, a dated phylogeny was constructed with the BEAST package (Bouckaert et al., 2014). Four independent runs of 100,000,000 MCMC generations, saving one tree in every 10,000 generations, were constructed in BEAUti 2.6.5 and implemented in BEAST 2.6.3 through the CIPRES Science Gateway (Miller et al., 2010). For each run, a lognormal relaxed clock and the birth–death tree prior were selected. bModelTest 1.1.2 (Bouckaert and Drummond, 2017), as implemented in BEAST, was used to determine the best-fit model for the COI dataset which was subsequently determined to be a variant of the HKY (with an additional group for the rates rct and rgt; 121,323). As fossil dating is challenging for Burnupia, the phylogeny was calibrated using published molecular clock rate for the COI gene in Wilke et al. (2009). The clock rate was set to the 95% confidence intervals found therein and linearly distributed [COI = 1.695% per million years; standard deviation (SD) = 1.33%–2.06%]. Thereafter, LogCombiner 2.5.2 (Bouckaert et al., 2014) was used to combine trees and log files of each run with 75% burn-in removed. Validation of the convergence and mixing of the combined log file was assessed in Tracer 1.7.1 (Rambaut et al., 2014) to ensure that all effective sample size (ESS) values were ≥200 and TreeAnnotator 2.5.2 (Bouckaert et al., 2014) was used to summarise trees, with no further burn-in removed.
Species Delimitation
Three species delimitation methods were followed to get an idea on the species richness and population structure in Burnupia in the Lesotho Highlands and surroundings: These included the General Mixed Yule-coalescent (GMYC), bayesian Poisson Tree Processes (bPTP) and Discriminant Analysis of Principal Components (DAPC). In all species delimitation instances, the outgroup (P. pileolus) was removed from sequence alignment files or input topologies (using TreeGraph 2; Stöver and Müller, 2010) to limit impacting Burnupia spp. delimitation results. The GMYC species delimitation method made use of the amended time-calibrated, ultrametric tree (generated in BEAST) and run using single and multiple thresholds, while the bPTP species delimitation method made use of the amended Bayesian topology (generated through MrBayes). All settings were otherwise kept as default for these approaches and the analyses were run through the online server at https://species.h-its.org/ptp/. The DAPC approach was performed using the adegenet 1.4-1 (Jombart and Bateman, 2008) package as run in the R statistical environment 4.0.2 (R Core Team, 2020) using a fasta alignment of all Burnupia haplotypes. Herein, Bayesian Information Criterion (BIC; Schwarz, 1978) was used to determine the optimal number of genetic clusters (k) before the DAPC analysis defined the specimens in each cluster.
Ancestral Distribution Analyses
To gain better perspectives of the evolutionary origins or colonisations of the established Burnupia phylospecies in the Lesotho Highlands ancestral area estimation was assessed across the dated phylogeny. BioGeoBEARS (Matzke, 2013) was implemented in RASP 4.2 (Yu et al., 2015) and used to estimate ancestral areas under six different biogeographical models for the entire dated phylogeny. This included the DEC, DIVA-like and BayArea-like models, including the +J parameter for each. Here, a simplified analysis with default settings in which only two areas were predefined (namely “within the Lesotho Highlands” and “outside the Lesotho Highlands”) was run and the best-fit model was determined by using the AIC—as implemented in BioGeoBEARS.
Isolation by Elevation and Distance
To assess the level of genetic diversity that can be attributed to Isolation by Distance (IBD) and Elevation (IBE), and thus help identify if the genus may contain cold-evolved species (i.e., those restricted by elevation and/or geographic distance at high altitudes), analyses of IBD and IBE were calculated within the genus as a whole following the methods of Sands et al. (2019b). In summary IBE and IBD was calculated using genetic, elevation and geographic distance matrices between samples (as compiled in GenAlEx 6.5; Peakall and Smouse, 2006) and following the distance-based redundancy analysis method (db-RDA; Legendre and Anderson, 1999; Legendre and Fortin, 2010) with the package vegan 2.5-4 (Oksanen et al., 2019) in the R statistical environment 4.0.2 (R Core Team, 2020).
ResultsPhylogenetics and Species Diversity
Between the BI and MP topologies generated in MrBayes and Paup respectively there are 19 supported nodes among the relationships of the 47 Burnupia specimens (PP ≥ 0.95 and/or BS ≥ 70; Figure 3). Species delimitation results range from between 10–16 Burnupia spp. among the dataset (GMYC = 16, bPTP = 15, DAPC = 10; Figure 3). The DAPC approach is the most conservative and, although not always reflective of phylogenetic support, best corresponds to the supported clade structures of the phylogenies. The other methods (i.e., GMYC and bPTP) tend to define species across multiple unsupported nodes creating increased complications with phylogenetic support. The ten species classified by the DAPC approach and regions where specimens of each were found are as follows: Burnupia sp. 1 (Lesotho Highlands), Burnupia sp. 2 (northern and western Lesotho Highlands and the South African Highveld), Burnupia sp. 3 (South African Highveld) and Burnupia sp. 4 (lower slopes of the eastern Drakensberg, KwaZulu-Natal), Burnupia sp. 5 (lower slopes of the south-eastern Drakensberg, KwaZulu-Natal down into the Eastern Cape), Burnupia sp. 6 (Tsitsikamma), Burnupia sp. 7 (Katberg Mountains), Burnupia sp. 8 (South African Lowveld and KwaZulu-Natal), Burnupia sp. 9 (Great Lake drainages of western Uganda) and Burnupia sp. 10 (Wild Coast) (Figure 3).
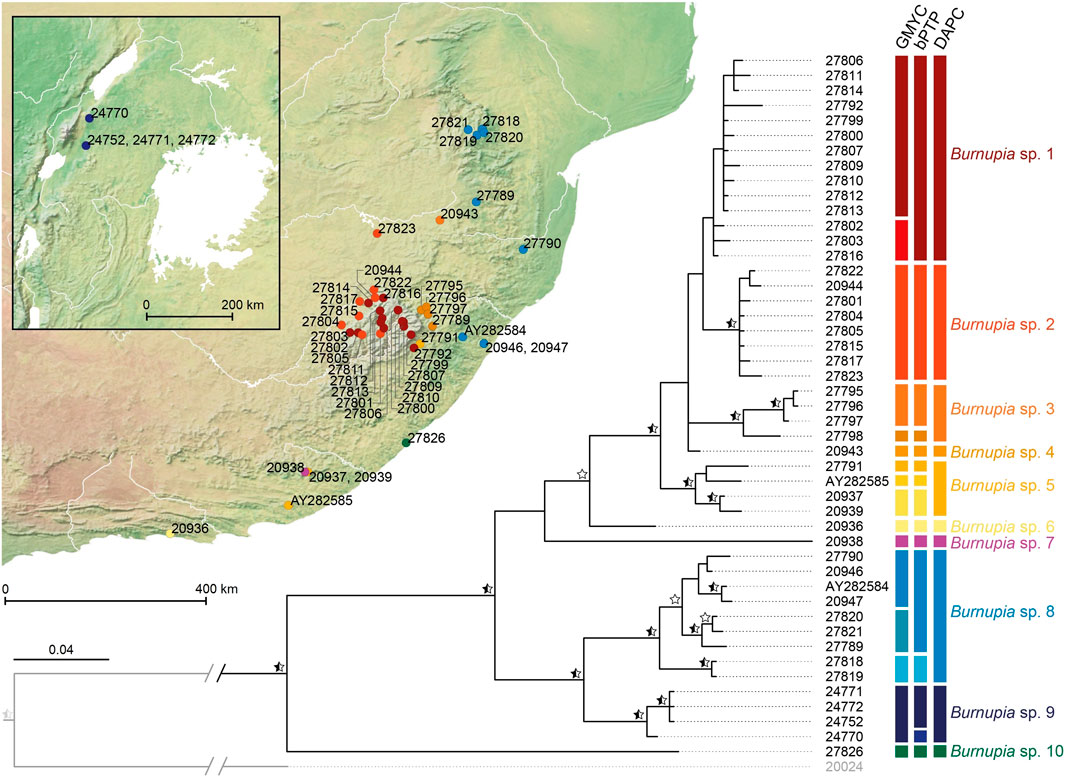
FIGURE 3. Bayesian phylogeny of Burnupia generated in MrBayes. Supported nodes (PP ≥ 0.95) are marked with black stars. Nodes additionally supported by parsimony analyses (Paup; BS ≥ 70) are marked with white stars or half black/white stars where PP support already exists. Results of the species delimitation methods (i.e., GMYC, bPTP and DAPC) are given alongside the tips in the form of coloured bars where different colours within each method denote different species and shared colours across methods indicate shared species. A distribution map with dots coloured and following the DAPC derived species are also presented. Further details on the samples and localities can be found in Table 1 following the specimen alphanumeric labels.
Dated Phylogeny and Ancestral Distributions
The genus Burnupia plausibly arouse somewhere between 25.95–5.64 Ma (data not displayed in Figure 4). While this study lacks several species of Burnupia (e.g., those from South America—which possibly represent a separate genus), the species occupying southern Africa likely began interspecific diversification from a common ancestor around 9.26 Ma [95% highest posterior density (HPD): 13.45–5.64 Ma; Figure 4A]. Following the DAPC species delamination results, by 0.60 Ma (95% HPD: 1.00–0.29 Ma; for Burnupia sp. 1 and Burnupia sp. 2) all species have diverged and the remaining genetic diversity is held intraspecifically (Figure 4A).
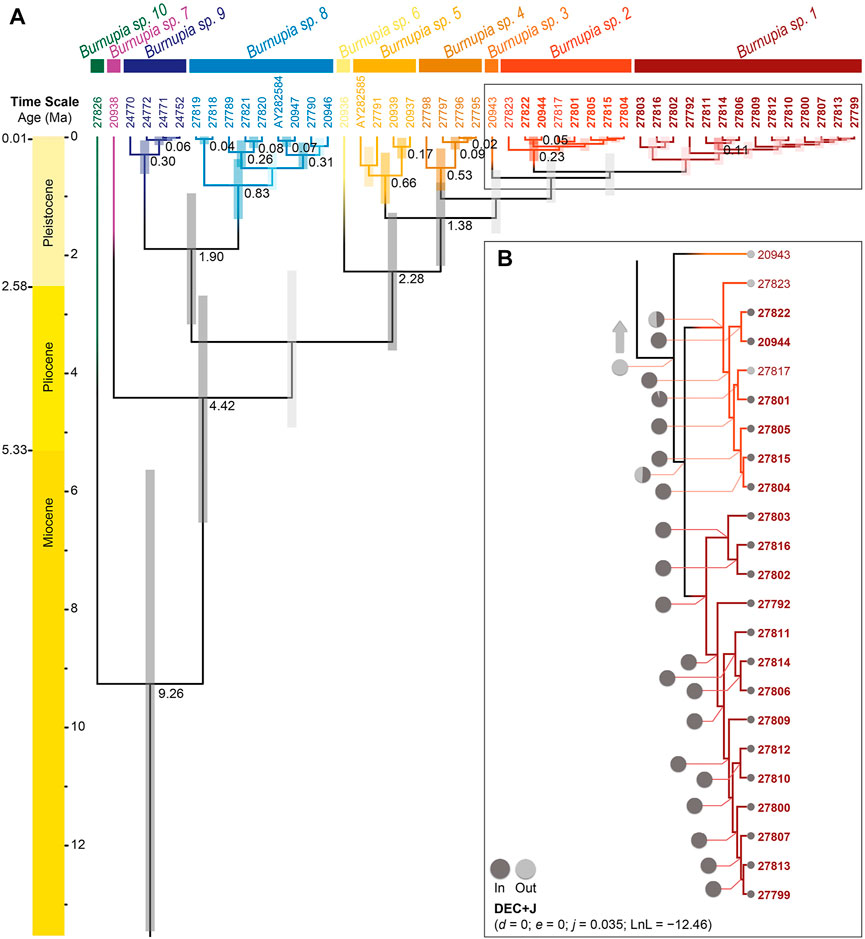
FIGURE 4. (A) The dated phylogeny of Burnupia spp. sampled across southern and eastern Africa constructed in BEAST based on COI sequence data. Supported divergence events (PP ≥ 0.95) are indicated by written dates at the nodes (in millions of years ago; Ma) and brighter shaded 95% credibility interval (i.e., HPD) bars. Species or clades are partitioned and coloured according to the DAPC species delimitation results (see Figure 2). Tip labels correspond to the preparation numbers of specimens noted in Table 1 and, therein, those in bold correspond to specimens occurring in the Lesotho Highlands. (B) The portion of the DEC+J results of the ancestral area estimation for Burnupia spp. on the dated phylogeny that correspond to Lesotho Highland occurrences. Ancestral area estimations were obtained from BioGeoBEARS run through RASP. Nodal pie charts represent the relative likelihood of the lineage occurring within the Lesotho Highlands (dark grey) and outside of the Lesotho Highlands (light grey). Smaller pies at the tips represent the current occurrence of each specimen. All other nodes across the phylogeny (data not shown; indicated by the light grey arrow) are equivalent to ancestral areas outside of the Lesotho Highlands.
Ancestral distribution estimations suggest that the common ancestors of all Burnupia species included in this study is not likely to have occurred in the Lesotho Highlands (Figure 4B). The analysis demonstrates only Burnupia sp. 1 and Burnupia sp. 2 have had historical presences in the Lesotho Highlands. The results point to the common ancestor of these two species likely colonising the Lesotho Highlands between 1.12–0.29 Ma, however significant PP and BS support for nodes surrounding this clade could not be found across our phylogenetic topologies (Figure 4A). Nevertheless, given species of Burnupia from the Lesotho Highlands seem to be closely related and Burnupia sp. 1, Burnupia sp. 2 and Burnupia sp. 3 diverged from Burnupia sp. 4 around 1.38 Ma (95% HPD: 2.19–0.78 Ma; PP ≥ 0.95 and BS ≥ 70; Figures 3, 4), this would indicate colonisation only occurred after this point (Figure 4).
Isolation by Distance and Elevation
Within the genus Burnupia as a whole, both IBD and IBE were significant yet both very weak in accounting for genetic diversity between species within the genus. We found R2 = 0.097 (p R2 = 0.102 (p Supplementary Material).
DiscussionDiversity and Biogeography in the Highlands
Although not particularly species rich in Burnupia, our results point to the Lesotho Highlands as being an important area for freshwater diversity.
Our results support between 10–16 phylogenetic species established through the molecular delimitation methods across, primarily southern Africa but also Uganda, albeit limited sampling the latter (Figures 2B, 3). Following the most conservative delimitation method, the DAPC approach, that best corresponds to our phylogenetic support and clade structure in our trees, two species can be found in the Lesotho Highlands, six can be found in the lower laying areas of KwaZulu-Natal and the Eastern Cape, three in the inland areas of South Africa (i.e., in the Highveld and Lowveld) and a single species was identified in the Western Region of Uganda (Figure 3). Diversity in the Lesotho Highlands is therefore to be seen as relatively average in comparison to at least the broader southern African region. However, given the temporal persistence of Burnupia in the Highlands and the endemicity of at least one of these species our results do suggest the region is important for cold-evolved freshwater biodiversity. Similar patterns have been observed for other aquatic organisms in the region (Daniels et al., 2003; Tolley et al., 2010). Our study is based on mitochondrial DNA variation. Studies in the European freshwater limpet genus Ancylus has demonstrated that diversity of young species can be underestimated—genome-wide approaches can help detecting hidden diversity in such cases (Weiss et al., 2018).
It is interesting that Burnupia partly overlaps with two other cold-evolved endemic species in the Highlands (Prinsloo and van Eeden, 1973). Bulinus sp. occurs up to 3,100 m.a.s.l. in Lesotho (Tumwebaze et al., 2022) and Galba mweruensis reaches similar altitudes (Mahulu et al., 2019). These two species are primarily found in stagnant and even temporary waters, whereas Burnupia species predominate on hard substrates as found in the permanent streams in the Highlands. A high oxygen demand and less drought tolerance has been shown for Burnupia (but see de Kock and Wolmarans, 2017) and some species have been used in lowland regions for ecotoxicological monitoring (e.g., Gerhardt and Palmer, 1998; Davies-Coleman and Palmer, 2004; Vellemu et al., 2018). Temperature sensitivity of Burnupia could potentially be established as a proxy for ongoing climatic changes, also in other Afromontane regions. In eastern Africa Burnupia also coincides with Bulinus and Galba, for example in the Ethiopian Highlands and the Aberdares Mountains, the Mau region and the Mount Kenya Massif (Brown, 1994; Mahulu et al., 2019; Tumwebaze et al., 2022). Comparative phylogeographical studies should be conducted to establish general patterns for freshwater benthic organisms in the Afromontane archipelago of sky-islands. Such studies could also help understanding better the means of colonisation across various taxa (Daniels et al., 2003; Tolley et al., 2010; Taylor et al., 2020). For Burnupia, the intrinsic characteristics of this limpet group, restrictive active dispersal and the narrow ecological tolerances question the actual dispersal and thus colonisation mechanism. Given that populations occur along basically whole drainage systems from lowland to sometimes up to the spring regions, jump dispersal is less likely. On the other hand, the nature of the habitat, high currents and the mobility of the (gravel) substrate restricts the ability of active dispersal upstream tremendously in aquatic mollusc (Kappes and Haase, 2012), more even for limpets (Albrecht et al., 2006). Birds as dispersal vector are probably not important in this case of upstream movement. Fish are often invoked as biological passive vector (Kappes and Haase, 2012). In fact, there are candidate species of fish including Labeobarbus yellow fishes (see Plug et al., 2010; Schrijvershof, 2015). Their role in the dispersal context of Burnupia should be studied in a comparative population-genetics framework.
The common ancestor of Burnupia sp. 1 and Burnupia sp. 2 was likely the first to colonise the region (Figure 4B). Our temporal reconstruction of the evolution of Burnupia suggests this may have occurred around 1.12–0.29 Ma, however phylogenetic support for the divergence events between Burnupia sp. 1, Burnupia sp. 2 and Burnupia sp. 3 are not well supported. Nevertheless, given the closest supported nodes, colonisation certainly happened post 1.38 Ma (95% HPD: 2.19–0.78 Ma; Figure 4) and prior to 0.23 Ma (95% HPD: 0.41–0.08 Ma; Figure 4). This Pleistocene period means Burnupia’s persistence in the Highlands likely overlapped with possibly several glacial maxima (Elderfield et al., 2012). Glacial cycles probably had two effects: firstly, glacial maxima caused the freezing of suitable freshwater environments in the Highlands (Mills et al., 2009, 2012; Hall and Meiklejohn, 2011)—this likely drove the several allopatric speciation events that can reasonably explain the speciation of Burnupia sp. 1 and Burnupia sp. 2 and the population structures therein. Secondly, glacial cycles probably drove several retreats and recolonisations (i.e., sinking) in Burnupia sp. 2 (Figure 4) and likely forced Burnupia sp. 1 to become cold-evolved given the endemicity of this species in the Highlands (Figures 3, 4), which likely accounts for at least some signals of IBD and IBE within Burnupia. Given our results, the Burnupia might serve as an ideal model group for testing the existence and extent of refugia and generally the role of climatic changes in rapidly changing Afromontane environments and colonisation patterns and processes, including co-evolution with dispersal vectors. As such, Burnupia could well become an Austral pendant to freshwater limpets in the northern hemisphere such as Ancylus (Albrecht et al., 2006) or Acroloxus (Stelbrink et al., 2016). An important asset in such studies would be an enhanced and better resolved taxonomic framework for all African Burnupia spp.
Taxonomic Implications
Freshwater scientists operating in Africa have long been advocating for a molecular based review of the genus Burnupia given the distribution of type localities among described species and shallow morphological differences among certain groups (Brown, 1994; Albrecht and Clewing, 2019), yet our molecular results only partly elucidate the complex picture. Brown (1994) presumed a very high level of synonymies among the available names but still listed no less than 21 species of Burnupia in his seminal book.
Our sampling regime largely overlapped the type localities of 14 of the 22 Burnupia species described from Africa as well as two subspecies of B. capensis (Figure 2). It was therefore interesting to find between 10–16 phylogenetic species established through the molecular delimitation methods (Figure 2). While the DAPC approach showed the best correspondence between the phylogenetically supported clade structure and the expected morphological diversity, the distribution of phylogenetic species did not entirely always correspond well with the distribution of described species and their type localities (Figures 2, 3). For example, at least six species have been described or noted in the KwaZulu-Natal Midlands (Figure 2A), yet through our sampling we could only find a single species in this area (i.e., Burnupia sp. 8; Figure 3). This species corresponds to B. caffra (Albrecht et al., 2004). While our sampling regime did not specifically target type localities, it is not unreasonable to suggest careful review and possible synonymisation of B. caffra, B. capensis natalensis, B. gordonensis, B. nana, B. obtusala and B. ponsonbyi may need to be looked at, especially given the broad distribution range of Burnupia sp. 8 (i.e., B. caffra) and that some of these described species share type localities in very close proximity to one another—sometimes even in the same drainage systems (Brown, 1994). Additionally, only a single species has been described (i.e., having a type locality) from the Eastern Cape region of South Africa (i.e., B. stenochorias; Figure 2A), yet we found three seemingly endemic lineages in this area (i.e., Burnupia sp. 6, Burnupia sp. 7 and Burnupia sp. 10; Figure 3). These could possibly represent the range extremities of Western Cape species whose type localities and systems were sampled but specimens failed to be amplified for COI (i.e., B. capensis capensis, B. gaulus and B. verreauxii), however, B. c. capensis and B. gaulus have been synonymised (see Brown, 1994) and it seems more realistic that they are as yet undescribed lineages given the lack of drainage collection between the southern Western Cape type localities and the large environmental and climatic shifts up the east coast of southern Africa (Werger, 1978; Abell et al., 2008; Linder et al., 2012). Placing names to phylogenetic species is therefore still a major challenge.
These naming uncertainties also extend to the Burnupia diversity in the Lesotho Highlands. For example, the distribution of Burnupia sp. 1 largely conforms with that attributed to B. capensis striatissima, while Burnupia sp. 2 conforms with that noted for B. trapezoidea (Brown, 1994; de Kock and Wolmarans, 2009, 2017). However, it seems unlikely that B. capensis striatissima is a subspecies of B. capensis given the endemicity in the Highlands and very close relationship with Burnupia sp. 2—noted as B. trapezoidea (see de Kock and Wolmarans, 2017; Figures 3, 4). The name B. trapezoidea (described from modern day Botswana) for Burnupia sp. 2 itself also requires careful review given three species (i.e., B. mooiensis, B. transvaalensis and B. vulcanus) have been described across the Highveld, particularly in the Vaal River catchments (Brown, 1994; Figures 2A, 3).
The taxonomic complications and conflicts in Burnupia are not unexpected. Other freshwater limpets have a similar history of splitting and lumping, which often masks real biodiversity (e.g., in Ancylus; Albrecht et al., 2006). Increasingly in aquatic malacology, when traditional shell-based taxonomic approaches are reviewed with molecular data and modern morphological methods, differences in shell shapes and patterns are noted to sometimes be poor tools for distinguishing species and likely to be influenced by environmental changes in some groups (Schultheiss et al., 2009; Sands et al., 2020; van Bocxlaer et al., 2021). Such changes have even been documented in a variety of molluscs in laboratory settings by small alterations in their environments (Neumann, 1959; Kistner and Dybdahl, 2013). If the same is corroborated for Burnupia, research involving type material is vitally needed to properly update the systematics of the genus and attribute the correct names to the phylogenetic species discovered.
Conclusion and Future Outlooks
Our study revealed that the Lesotho Highlands are an important region for freshwater diversity and likely holds cold-evolved species. These Highlands were colonised roughly during Calabrian–Chibanian stages of the Pleistocene by Burnupia species and may be a useful model to study the evolution of the Highlands and likewise its impact on freshwater organisms given indications of population structure. It requires NGS approaches to be used which are more sensitive to reconstructing population‐level history. The study has improved our knowledge of Burnupia systematics, but it still remains mostly unresolved. Further research is desperately needed using type material and establishing if morphological features such as shell shape and patterning is affected by environmental conditions.
Data Availability Statement
The DNA sequence data presented in this study can be found in the NCBI’s GenBank repository online (www.ncbi.nlm.nih.gov/genbank). The accession numbers can be found in Table 1.
Ethics Statement
Ethical review and approval was not required for the study. All material was sampled through bilateral agreements among collaborating institutes or with local permits where required.
Author Contributions
The team of authors has a strong research focus on freshwater biogeography, phylogenetics and geology—especially in Africa. All authors contributed to the general framework of the article: CA, VSG, and FR performed fieldwork and collected samples; CA generated the data; AFS performed the analyses; AFS and CA led the writing.
Funding
The research herein forms part of a broader Lesotho project which received funding from the DFG (AL 1076/12-1 and RI 809/38-1).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We wish to kindly thank C. Clewing, U. Bößneck, J. Day and J. Tumusiime for their assistance in the field or for their help providing samples (South Africa, Lesotho, Uganda). Moreover, we, the authors, are particularly grateful to the reviewers and editor for their valuable appraisals and suggestions during the publication process.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2022.914272/full#supplementary-material
References
Abell, R., Thieme, M. L., Revenga, C., Bryer, M., Kottelat, M., Bogutskaya, N., et al. (2008). Freshwater Ecoregions of the World: A New Map of Biogeographic Units for Freshwater Biodiversity Conservation. BioScience 58, 403–414. doi:10.1641/b580507
CrossRef Full Text | Google Scholar
Akaike, H. (1973). “Information Theory and an Extension of the Maximum Likelihood Principle,” in Proceedings of the Second International Symposium on Information Theory. Editors B. N. Petrov, and B. F. Csaki (Budapest: Academiai Kiado), 267–281.
Google Scholar
Albrecht, C., and Clewing, C. (20192017). “Burnupiidae Albrecht,” in Freshwater Mollusks of the World. Editors C. Lydeard, and K. S. Cummings (Baltimore, USA: Johns Hopkins University Press), 172–174.
Google Scholar
Albrecht, C., Trajanovski, S., Kuhn, K., Streit, B., and Wilke, T. (2006). Rapid Evolution of an Ancient Lake Species Flock: Freshwater Limpets (Gastropoda: Ancylidae) in the Balkan Lake Ohrid. Org. Divers. Evol. 6, 294–307. doi:10.1016/j.ode.2005.12.003
CrossRef Full Text | Google Scholar
Albrecht, C., Wilke, T., Kuhn, K., and Streit, B. (2004). Convergent Evolution of Shell Shape in Freshwater Limpets: The African Genus Burnupia. Zool. J. Linn. Soc-Lond. 140, 577–586. doi:10.1111/j.1096-3642.2003.00108.x
CrossRef Full Text | Google Scholar
Archer, E., Engelbrecht, F., Hänsler, A., Landman, W., Tadross, M., and Helmschrot, J. (2018). Seasonal Prediction and Regional Climate Projections for Southern Africa. Biodivers. Ecol. 6, 14–21. doi:10.7809/b-e.00296
CrossRef Full Text | Google Scholar
Bentley, L. K., Robertson, M. P., and Barker, N. P. (2018). Range Contraction to a Higher Elevation: The Likely Future of the Montane Vegetation in South Africa and Lesotho. Biodivers. Conserv. 28, 131–153. doi:10.1007/s10531-018-1643-6
CrossRef Full Text | Google Scholar
Bouckaert, R., Heled, J., Kühnert, D., Vaughan, T., Wu, C.-H., Xie, D., et al. (2014). BEAST 2: A Software Platform for Bayesian Evolutionary Analysis. PLOS Comput. Biol. 10, e1003537. doi:10.1371/JOURNAL.PCBI.1003537
PubMed Abstract | CrossRef Full Text | Google Scholar
Brighenti, S., Hotaling, S., Finn, D. S., Fountain, A. G., Hayashi, M., Herbst, D., et al. (2021). Rock Glaciers and Related Cold Rocky Landforms: Overlooked Climate Refugia for Mountain Biodiversity. Glob. Change Biol. 27, 1504–1517. doi:10.1111/gcb.15510
CrossRef Full Text | Google Scholar
Brown, D. S. (1994). Freshwater Snails of Africa and Their Medical Importance. London: Taylor & Francis.
Google Scholar
Chatanga, P., and Seleteng-Kose, L. (2021). Montane Palustrine Wetlands of Lesotho: Vegetation, Ecosystem Services, Current Status, Threats and Conservation. Wetlands 41, 67. doi:10.1007/s13157-021-01470-1
CrossRef Full Text | Google Scholar
Clewing, C., Geertz, T., Rassam, H., Woldekiros, T. H., and Albrecht, C. (2022). Freshwater Diversity at a Biogeographic Edge Zone: The High-Mountain Pea-Clams of Ethiopia. Syst. Biodivers. 20, 1–15. doi:10.1080/14772000.2021.2005706
CrossRef Full Text | Google Scholar
Connolly, M. K. W. (1939). A Monographic Survey of the South African Non-Marine Mollusca. Ann. Natal. Mus. 33, 1–660.
Google Scholar
Dallas, H. F., and Rivers-Moore, N. (2014). Ecological Consequences of Global Climate Change for Freshwater Ecosystems in South Africa. S. Afr. J. Sci. 110, 1–11. doi:10.1590/sajs.2014/20130274
CrossRef Full Text | Google Scholar
Daniels, S. R., Gouws, G., Stewart, B. A., and Coke, M. (2003). Molecular and Morphometric Data Demonstrate the Presence of Cryptic Lineages Among Freshwater Crabs (Decapoda: Potamonautidae: Potamonautes) From the Drakensberg Mountains, South Africa. Biol. J. Linn. Soc. 78, 129–147. doi:10.1046/j.1095-8312.2003.00143.x
CrossRef Full Text | Google Scholar
Davies-Coleman, H. D., and Palmer, C. G. (2004). “The Use of a Freshwater Mollusc, Burnupia stenochorias (Ancylidae) as an Ecotoxicological Indicator in Whole Effluent Toxicity Testing,” in Proceedings of the Water Institute of Southern Africa (WISA) Biennial Conference (Cape Town: Document Transformation Technologies), 309–315.
Google Scholar
Davies-Coleman, H. D. (2001). The Growth and Reproduction of the Freshwater Limpet Burnupia stenochorias (Pulmonata, Ancylidae), and an Evaluation of its Use as an Ecotoxicology Indicator in Whole Effluent Testing. Grahamstown (EC): Rhodes University. [doctoral dissertation].
Google Scholar
de Kock, K. N., and Wolmarans, C. T. (2016). Distribution and Habitats of Burnupia mooiensis in South Africa: (Walker 1912 [Gastropoda: Ancylidae]). S. Afr. J. Sci. Technol. 35, a1372. doi:10.4102/satnt.v35i1.1372
CrossRef Full Text | Google Scholar
de Kock, K. N., and Wolmarans, C. T. (2017). Distribution and Habitats of Burnupia trapezoidea (Boettger, 1910) (Gastropoda: Ancylidae) in South Africa. Water SA 43, 258–263. doi:10.4314/wsa.v43i2.09
CrossRef Full Text | Google Scholar
de Kock, K., and Wolmarans, C. (2009). Distribution of Burnupia capensis (Walker, 1912) and Burnupia stenochorias (Melvill & Ponsonby, 1903) (Gastropoda: Ancylidae) in South Africa. S. Afr. J. Sci. Technol. 28, 220–236. doi:10.4102/satnt.v28i3.59
CrossRef Full Text | Google Scholar
Elderfield, H., Ferretti, P., Greaves, M., Crowhurst, S., McCave, I. N., Hodell, D., et al. (2012). Evolution of Ocean Temperature and Ice Volume Through the Mid-Pleistocene Climate Transition. Science 337, 704–709. doi:10.1126/science.1221294
PubMed Abstract | CrossRef Full Text | Google Scholar
Eriksson, P. G. (1981). A Palaeoenvironmental Analyses of the Clarens Formationin the Natal Drakensberg. Trans. Geol. S. Afr. 84, 7–17.
Google Scholar
Folmer, O., Black, M., Hoeh, W., Lutz, R., and Vrijenhoek, R. (1994). DNA Primers for Amplification of Mitochondrial Cytochrome C Oxidase Subunit I from Diverse Metazoan Invertebrates. Mol. Mar. Biol. Biotechnol. 3, 294–299.
PubMed Abstract | Google Scholar
Füreder, L., Ettinger, R., Boggero, A., Thaler, B., and Thies, H. (2006). Macroinvertebrate Diversity in Alpine Lakes: Effects of Altitude and Catchment Properties. Hydrobiologia 562, 123–144. doi:10.1007/s10750-005-1808-7
CrossRef Full Text | Google Scholar
Gerhardt, A., and Palmer, C. (1998). Copper Tolerances of Adenophlebia auriculata (Eaton) 1884 (Insecta: Ephemeroptera) and Burnupia stenochorias Cawston 1932 (Gastropoda: Ancylidae) in Indoor Artificial Streams. Sci. Total Environ. 215, 217–229. doi:10.1016/s0048-9697(98)00121-1
CrossRef Full Text | Google Scholar
Hågvar, S., Gobbi, M., Kaufmann, R., Ingimarsdóttir, M., Caccianiga, M., Valle, B., et al. (2020). Ecosystem Birth Near Melting Glaciers: A Review on the Pioneer Role of Ground-Dwelling Arthropods. Insects 11, 644. doi:10.3390/INSECTS11090644
CrossRef Full Text | Google Scholar
Hall, K., and Meiklejohn, I. (2011). “Glaciation in Southern Africa and in the Sub-Antarctic,” in Developments in Quaternary Science: Quaternary Glaciations – Extent and Chronology. Editors J. Ehlers, P. L. Gibbard, and P. H. Hughes (Amsterdam: Elsevier), 1081–1085. doi:10.1016/b978-0-444-53447-7.00078-7
CrossRef Full Text | Google Scholar
Hoogendoorn, G., Stockigt, L., Saarinen, J., and Fitchett, J. M. (2020). Adapting to Climate Change: The Case of Snow-Based Tourism in Afriski, Lesotho. Afr. Geogr. Rev. 40, 92–104. doi:10.1080/19376812.2020.1773878
CrossRef Full Text | Google Scholar
Kappes, H., and Haase, P. (2012). Slow, but Steady: Dispersal of Freshwater Molluscs. Aquat. Sci. 74, 1–14. doi:10.1007/s00027-011-0187-6
CrossRef Full Text | Google Scholar
Kistner, E. J., and Dybdahl, M. F. (2013). Adaptive Responses and Invasion: The Role of Plasticity and Evolution in Snail Shell Morphology. Ecol. Evol. 3, 424–436. doi:10.1002/ece3.471
PubMed Abstract | CrossRef Full Text | Google Scholar
Kopij, G. (2015). Avian Assemblages in Afro-Mountain and Alti-Mountain Grasslands in the Endemic Maloti/Drakensberg Region of Lesotho. Zoology Ecol. 25, 319–326. doi:10.1080/21658005.2015.1095854
CrossRef Full Text | Google Scholar
Legendre, P., and Anderson, M. J. (1999). Distance-Based Redundancy Analysis: Testing Multispecies Responses in Multifactorial Ecological Experiments. Ecol. Monogr. 69, 1–24. doi:10.1890/0012-9615(1999)069[0001:dbratm]2.0.co;2
CrossRef Full Text | Google Scholar
Legendre, P., and Fortin, M.-J. (2010). Comparison of the Mantel Test and Alternative Approaches for Detecting Complex Multivariate Relationships in the Spatial Analysis of Genetic Data. Mol. Ecol. Resour. 10, 831–844. doi:10.1111/j.1755-0998.2010.02866.x
PubMed Abstract | CrossRef Full Text | Google Scholar
Linder, H. P., de Klerk, H. M., Born, J., Burgess, N. D., Fjeldså, J., and Rahbek, C. (2012). The Partitioning of Africa: Statistically Defined Biogeographical Regions in Sub-Saharan Africa. J. Biogeogr. 39, 1189–1205. doi:10.1111/j.1365-2699.2012.02728.x
CrossRef Full Text | Google Scholar
Mahulu, A., Clewing, C., Stelbrink, B., Chibwana, F. D., Tumwebaze, I., Russell Stothard, J., et al. (2019). Cryptic Intermediate Snail Host of the Liver Fluke Fasciola hepatica in Africa. Parasit. Vectors 12, 573–611. doi:10.1186/s13071-019-3825-9
PubMed Abstract | CrossRef Full Text | Google Scholar
Matzke, N. J. (2013). Probabilistic Historical Biogeography: New Models for Founder-Event Speciation, Imperfect Detection, and Fossils Allow Improved Accuracy and Model-Testing. [Berkeley (CA)]: University of California. [doctoral dissertation].
CrossRef Full Text | Google Scholar
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees,” in 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14-14 Nov. 2010 (IEEE). doi:10.1109/GCE.2010.5676129
CrossRef Full Text | Google Scholar
Mills, S. C., Grab, S. W., and Carr, S. J. (2009). Recognition and Palaeoclimatic Implications of Late Quaternary Niche Glaciation in Eastern Lesotho. J. Quat. Sci. 24, 647–663. doi:10.1002/jqs.1247
CrossRef Full Text | Google Scholar
Mills, S. C., Grab, S. W., Rea, B. R., Carr, S. J., and Farrow, A. (2012). Shifting Westerlies and Precipitation Patterns during the Late Pleistocene in Southern Africa Determined Using Glacier Reconstruction and Mass Balance Modelling. Quat. Sci. Rev. 55, 145–159. doi:10.1016/j.quascirev.2012.08.012
CrossRef Full Text | Google Scholar
Neumann, D. (1959). Morphologische und experimentelle Untersuchungen über die Variabilität Per Farbmuster auf der Schale von Theodoxus fluviatilis L. Z. Morph. U. Okol. Tiere 48, 349–411. doi:10.1007/bf00408578
CrossRef Full Text | Google Scholar
Norström, E., Bringensparr, C., Fitchett, J. M., Grab, S. W., Rydberg, J., and Kylander, M. (2018). Late-Holocene Climate and Vegetation Dynamics in Eastern Lesotho Highlands. Holocene 28, 1483–1494. doi:10.1177/0959683618777054
CrossRef Full Text | Google Scholar
Oksanen, J., Blanchet, G., Friendly, M., Kindt, R., Legendre, P., and McGlinn, D. (2019). Vegan: Community Ecology R Package. Available Online at: https://cran.r-project.org/package=vegan (accessed April 6, 2022).
Google Scholar
Peakall, R., and Smouse, P. E. (2006). GenAlEx 6: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research. Mol. Ecol. Notes 6, 288–295. doi:10.1111/j.1471-8286.2005.01155.x
CrossRef Full Text | Google Scholar
Perera, S. J., Herbert, D. G., Procheş, Ş., and Ramdhani, S. (2021). Land Snail Biogeography and Endemism in South-Eastern Africa: Implications for the Maputaland-Pondoland-Albany Biodiversity Hotspot. PLOS One 16, e0248040. doi:10.1371/journal.pone.0248040
PubMed Abstract | CrossRef Full Text | Google Scholar
Phiri, E. E., Daniels, S. R., Phiri, E. E., and Daniels, S. R. (2016). Multilocus Coalescent Species Delimitation Reveals Widespread Cryptic Differentiation Among Drakensberg Mountain-Living Freshwater Crabs (Decapoda: Potamonautes). Invert. Syst. 30, 60–74. doi:10.1071/is15035
CrossRef Full Text | Google Scholar
Plug, I., Mitchell, P., and Bailey, G. (2010). Late Holocene Fishing Strategies in Southern Africa as Seen from Likoaeng, Highland Lesotho. J. Archaeol. Sci. 37, 3111–3123. doi:10.1016/j.jas.2010.07.012
CrossRef Full Text | Google Scholar
Prinsloo, J. F., and van Eeden, J. A. (1973). The Distribution of the Freshwater Molluscs in Lesotho with Particular Reference to the Intermediate Host of Fasciola hepatica. Wetensk. Bydr. Potchefstroom Univ. B 57, 1–11.
Google Scholar
Pullanikkatil, D., and Urama, K. C. (2011). The Effects of Industrialisation on Water Quality and Livelihoods in Lesotho. Int. J. Environ. Eng. 3, 175–191. doi:10.1504/ijee.2011.039453
CrossRef Full Text | Google Scholar
Rahbek, C., Borregaard, M. K., Colwell, R. K., Dalsgaard, B., Holt, B. G., Morueta-Holme, N., et al. (2019). Humboldt’s Enigma: What Causes Global Patterns of Mountain Biodiversity? Science 365, 1108–1113. doi:10.1126/science.aax0149
PubMed Abstract | CrossRef Full Text | Google Scholar
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 61, 539–542. doi:10.1093/sysbio/sys029
PubMed Abstract | CrossRef Full Text | Google Scholar
Sands, A. F., Glöer, P., Guörlek, M. E., Albrecht, C., and Neubauer, T. A. (2020). A Revision of the Extant Species of Theodoxus (Gastropoda, Neritidae) in Asia, with the Description of Three New Species. Zoosyst. Evol. 96, 25–66. doi:10.3897/zse.96.48312
CrossRef Full Text | Google Scholar
Sands, A. F., Neubauer, T. A., Nasibi, S., Harandi, M. F., Anistratenko, V. V., Wilke, T., et al. (2019a). Old Lake versus Young Taxa: A Comparative Phylogeographic Perspective on the Evolution of Caspian Sea Gastropods (Neritidae: Theodoxus). R. Soc. Open Sci. 6, 190965. doi:10.1098/RSOS.190965
PubMed Abstract | CrossRef Full Text | Google Scholar
Sands, A. F., Sereda, S. v., Stelbrink, B., Neubauer, T. A., Lazarev, S., Wilke, T., et al. (2019b). Contributions of Biogeographical Functions to Species Accumulation May Change Over Time in Refugial Regions. J. Biogeogr. 46, 1274–1286. doi:10.1111/jbi.13590
CrossRef Full Text | Google Scholar
Schrijvershof, J. (2015). Biology and Ecology of Fishes of the Senqu River, Lesotho. Potchefstroom (NWP): North-West University. [Mater Dissertation].
Google Scholar
Schultheiss, R., van Bocxlaer, B., Wilke, T., and Albrecht, C. (2009). Old Fossils-Young Species: Evolutionary History of an Endemic Gastropod Assemblage in Lake Malawi. Proc. Biol. Sci. 276, 2837–2846. doi:10.1098/rspb.2009.0467
PubMed Abstract | CrossRef Full Text | Google Scholar
Serdeczny, O., Adams, S., Baarsch, F., Coumou, D., Robinson, A., Hare, W., et al. (2016). Climate Change Impacts in Sub-Saharan Africa: From Physical Changes to Their Social Repercussions. Reg. Environ. Change 17, 1585–1600. doi:10.1007/s10113-015-0910-2
CrossRef Full Text | Google Scholar
Skelton, P. (2000). Flagships and Fragments—Perspectives on the Conservation of Freshwater Fishes in Southern Africa. Afr. J. Aquatic Sci. 25, 37–42. doi:10.2989/160859100780177929
CrossRef Full Text | Google Scholar
Smith, R. M. H., Eriksson, P. G., and Botha, W. J. (1993). A Review of the Stratigraphy and Sedimentary Environments of the Karoo-Aged Basins of Southern Africa. J. Afr. Earth Sci. 16, 143–169. doi:10.1016/0899-5362(93)90164-l
CrossRef Full Text | Google Scholar
Stelbrink, B., Shirokaya, A. A., Föller, K., Wilke, T., and Albrecht, C. (2016). Origin and Diversification of Lake Ohrid’s Endemic Acroloxid Limpets: The Role of Geography and Ecology. BMC Evol. Biol. 16, 273–313. doi:10.1186/s12862-016-0826-6
PubMed Abstract | CrossRef Full Text | Google Scholar
Stöver, B. C., and Müller, K. F. (2010). TreeGraph 2: Combining and Visualizing Evidence from Different Phylogenetic Analyses. BMC Bioinformatics 11, 1–9. doi:10.1186/1471-2105-11-7
PubMed Abstract | CrossRef Full Text | Google Scholar
Swartz, E. R. (2005). Phylogeography, Phylogenetics and Evolution of the Redfins (Teleostei, Cyprinidae, Pseudobarbus) in Southern Africa. Pretoria (GP): University of Pretoria. [Doctoral Dissertation].
Google Scholar
Swartz, E. R., Skelton, P. H., and Bloomer, P. (2009). Phylogeny and Biogeography of the Genus Pseudobarbus (Cyprinidae): Shedding Light on the Drainage History of Rivers Associated with the Cape Floristic Region. Mol. Phylogenet. Evol. 51, 75–84. doi:10.1016/j.ympev.2008.10.017
PubMed Abstract | CrossRef Full Text | Google Scholar
Swofford, D. L. (2002). Phylogenetic Analysis Using Parsimony PAUP* 4.0 Beta Version Disclaimer and User Agreement. Sunderland, MA: Sinauer Associates.
Google Scholar
Taylor, C. L., Barker, N. P., Barber-James, H. M., Villet, M. H., and Pereira-da-Conceicoa, L. L. (2020). Habitat Requirements Affect Genetic Variation in Three Species of Mayfly (Ephemeroptera, Baetidae) From South Africa. ZooKeys 936, 1–24. doi:10.3897/zookeys.936.38587
PubMed Abstract | CrossRef Full Text | Google Scholar
Tolley, K. A., Braae, A., and Cunningham, M. (2010). Phylogeography of the Clicking Stream Frog Strongylopus grayii (Anura, Pyxicephalidae) Reveals Cryptic Divergence Across Climatic Zones in an Abundant and Widespread Taxon. Afr. J. Herpetol. 59, 17–32. doi:10.1080/04416651003744943
CrossRef Full Text | Google Scholar
Tumwebaze, I., Clewing, C., Chibwana, F. D., Kipyegon, J. K., and Albrecht, C. (2022). Evolution and Biogeography of Freshwater Snails of the Genus Bulinus (Gastropoda) in Afromontane Extreme Environments. Front. Environ. Sci. 10, 902900. doi:10.3389/fenvs.2022.902900
CrossRef Full Text | Google Scholar
Turpie, J., Benn, G., Thompson, M., and Barker, N. (2021). Accounting for Land Cover Changes and Degradation in the Katse and Mohale Dam Catchments of the Lesotho Highlands. Afr. J. Range For. Sci. 38, 53–66. doi:10.2989/10220119.2020.1846214
CrossRef Full Text | Google Scholar
van Bocxlaer, B., Clewing, C., Duputié, A., Roux, C., and Albrecht, C. (2021). Population Collapse in Viviparid Gastropods of the Lake Victoria Ecoregion Started Before the Last Glacial Maximum. Mol. Ecol. 30, 364–378. doi:10.1111/mec.15599
PubMed Abstract | CrossRef Full Text | Google Scholar
Vellemu, E., Mensah, P., Griffin, N., and Odume, O. (2018). Derivation of Scenario-Specific Water Quality Guidelines for Acid Mine Drainage in South Africa, Using a Risk-Based Approach. Afr. J. Aquat. Sci. 43, 51–58. doi:10.2989/16085914.2018.1446126
CrossRef Full Text | Google Scholar
Weber, T., Haensler, A., Rechid, D., Pfeifer, S., Eggert, B., and Jacob, D. (2018). Analyzing Regional Climate Change in Africa in a 1.5, 2, and 3°C Global Warming World. Earth’s Future 6, 643–655. doi:10.1002/2017ef000714
CrossRef Full Text | Google Scholar
Weiss, M., Weigand, H., Weigand, A. M., and Leese, F. (2018). Genome-Wide Single-Nucleotide Polymorphism Data Reveal Cryptic Species Within Cryptic Freshwater Snail Species—The Case of the Ancylus fluviatilis Species Complex. Ecol. Evol. 8, 1063–1072. doi:10.1002/ece3.3706
PubMed Abstract | CrossRef Full Text | Google Scholar
Werger, M. J. A. (1978). “Biogeographical Division of Southern Africa,” in Biogeography and Ecology of Southern Africa. Monographiae Biologicae. Editor M. J. A. Werger (Dordrecht: Springer), 145–170. doi:10.1007/978-94-009-9951-0_7
CrossRef Full Text | Google Scholar
Wilke, T., and Davis, G. M. (2000). Infraspecific Mitochondrial Sequence Diversity in Hydrobia ulvae and Hydrobia ventrosa (Hydrobiidae: Rissooidea: Gastropoda): Do Their Different Life Histories Affect Biogeographic Patterns and Gene Flow? Biol. J. Linn. Soc. 70, 89–105. doi:10.1111/j.1095-8312.2000.tb00202.x
CrossRef Full Text | Google Scholar
Wilke, T., Schultheiß, R., and Albrecht, C. (2009). As Time Goes By: A Simple Fool’s Guide to Molecular Clock Approaches in Invertebrates. Am. Malacol. Bull. 27, 25–45. doi:10.4003/006.027.0203
CrossRef Full Text | Google Scholar
Yu, Y., Harris, A. J., Blair, C., and He, X. (2015). RASP (Reconstruct Ancestral State in Phylogenies): A Tool for Historical Biogeography. Mol. Phylogenet. Evol. 87, 46–49. doi:10.1016/j.ympev.2015.03.008
PubMed Abstract | CrossRef Full Text | Google Scholar
Source link : https://www.frontiersin.org/articles/10.3389/fenvs.2022.914272
Author :
Publish date : 2022-05-11 07:00:00
Copyright for syndicated content belongs to the linked Source.





