Description of the study area
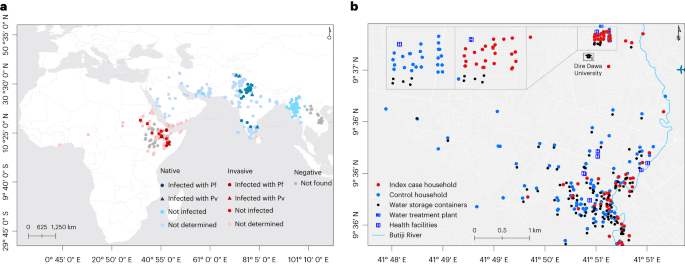
Dire Dawa, located 515 km southeast of Addis Ababa (capital of Ethiopia) and 311 km west of Djibouti, is a logistics hub for transportation of goods and cargo (Fig. 1b). Of its total population (445,050), 74% live in an urban area which is only 2.3% of the 1,288 km2 Dire Dawa City administrative land (UN-HABITAT, 2008). The area has a warm and dry climate with low level of precipitation (annual average rainfall of 624 mm), and an annual temperature ranging from 19 °C to 32 °C. Malaria incidence has historically been low (an annual parasite clinical incidence of P. falciparum and P. vivax infections.
We obtained public health data, collected through the District Health Information System 2 (DHIS2), to analyze the trend in malaria cases between 2015 and 2022. In the Ethiopian malaria case management guideline, microscopy is recommended for diagnosis at the health center level and above. RDTs are recommended to be used only at the health post level by community health extension workers, in rural settings. In all of the facilities located in Dire Dawa, microscopy was used for diagnosis. The DHIS2 data do not capture cases detected at private health facilities. The recent ‘Global framework for the response to malaria in urban areas’ by the WHO4 states that “In some urban settings, the private sector is a major source of malaria diagnosis and treatment. However, it is poorly integrated into the surveillance system”. To give context on how much is being managed by the private sector in Dire Dawa, we have collected 4 years of data (January 2019 to May 2022) from 34 out 39 health facilities (both private and public) that are located within the city administration. This included 2 public and 5 private hospitals, 15 health centers (funded publicly) and 17 clinics (private). Some private clinics (n = 5) refused to provide data or provided incomplete data. Goro Health Center and DDU students’ clinic were selected for the current study based on the highest number of cases they reported before the start of the study (January–February 2022). In fact, together, the two health facilities reported 56% of the total cases in the city in 2022 (January–May). As in all public universities in Ethiopia, students live within campus with full and shared accommodation provided by the government. At DDU, an average of six students of the same sex and year of study share a dormitory on a three-story building that has an average of 67 dormitories. Routine healthcare service is provided in a university dedicated students’ clinic.
Study design and procedure
To ascertain the effect of exposure to An. stephensi on malaria, we employed a case–control study where identification of patients was done prospectively to capture co-occurrent characteristics in terms of exposure and risk factors. We recruited consecutive patients with criteria described below in a 1:2 ratio (one case:two controls) unmatched study design. Study protocol was approved by the Institutional Ethical Review Board of Armauer Hansen Research Institute (AHRI)/All Africa Leprosy Education, Research, and Training Center ethics review committee (AF-10-015.1, PO/07/19). We obtained informed written consent from all participants and guardians or parents for minors.
Recruitment of participants
Patients with (history within 48 h) fever that presented at the two health facilities and tested positive for malaria by microscopy were recruited as index cases (index) from April to July 2022. We recruited febrile patients who attended the same clinic and tested negative for malaria as controls within 72 h of when the index was identified. The index and controls were followed to their homes, and their household/dormitory members were tested for malaria and their households/dormitories were screened for Anopheles mosquitoes (larvae and adult). Household/dormitory members of cases and controls who were willing to participate in the reactive case detection were included irrespective of their symptoms. Households were surveyed for mosquitoes when the head of the household and members of the dormitory gave consent to allow the study team to use mosquito collection methods in their houses/dormitories. Families of cases or controls who were not available within 72 h of recruitment of the cases or controls irrespective of their symptoms were excluded as well as individuals or family members who were unwilling/refused to give informed written consent. It is noticeable that, although the study was unmatched due to the difficulty in recruiting matched controls in geographical proximity of the cases, their general characteristics were very similar. Detailed characteristics of study participants are presented in Table 1.
Sample size
We planned an unmatched case:control ratio of approximately 1:2 (ref. 56) with prospective case identification until the stopping rule was achieved. The choice of the case:control ratio was based on a logistic regression model aimed to detect an OR of at least 2, assuming an exposure of 20% in controls at household level, where the exposure was defined as presence of An. stephensi. The power analysis was conducted in epiR package (R-cran software), and the stopping rule was set to a power of 70% for the study to be sufficiently powered to detect differences between the presence of malaria on An. stephensi exposure at household level. The controls were selected from the same population as the cases and post-stratification applied. Data from cases and controls were reviewed regularly, and final sample size was set to 290 with 101 cases and 189 controls. The recruitment of case household and control household members was done to include reactive case detection and improve the power of the study (as well as the OR minimum detection).
Data collection
Data on the sociodemographic, epidemiological, intervention and travel history were collected verbally using pre-tested questionnaires which were uploaded to mobile tablets using REDCap tools. The entomological survey data and intervention availability were scored by the study data collectors. Malaria case incidence data (from January 2019 to May 2022) were collected from the records of both private and public health facilities (n = 34).
Blood samples collection
Finger prick blood samples (~0.5 ml), collected in BD K2EDTA Microtainer tubes, were used to diagnose malaria using RDT (Abbott Bioline Malaria Ag Pf/Pv HRP2/LDH) and microscopy, and to prepare dried blood spots (DBS) on 3MM Whatman filter paper (Whatman). The remaining blood was separated into cell pellet and plasma. Slide films were confirmed by expert microscopists. Sociodemographic, epidemiological, intervention utilization, and history of travel and malaria were collected from all study participants.
Entomological surveys
We surveyed immature stages of Anopheles mosquitoes within a 100-m radius of the index and control houses/dormitories targeting both manmade water storage containers and natural habitats including riverbeds and stream edges. We checked each aquatic habitat for 10 min from 9:00 to 11:00 and 15:00 to 17:00 for the presence of Anopheles mosquitoes’ larvae or pupae aiming for ten dips per habitat (using a standard dipper with 350 ml capacity). Characteristics of water holding containers (permanency of habitat, lid status, purpose, volume, presence of shade, type, turbidity, temperature and water source) were recorded for each habitat (Extended Data Table 6). We searched adult mosquitoes using Prokopack aspirators for 10 min between 6:00 and 8:00 indoor, outdoor and in animal shelters located within the compound of the household or inside and outside the dormitories at the university (Extended Data Table 7). Mosquito surveys (immature and adult) were done within 48–72 h of when the index/control was recruited.
Conventional adult mosquito collection methods such as Centers for Disease Control and Prevention light traps and pyrethrum spray sheet have limited sensitivity for this invasive species mainly related with its unique resting behavior21. To supplement the evidence generated from the case–control study and examine the resting sites of the adult Anopheles mosquitoes in detail in the study area, additional adult mosquito surveys were done targeting potential resting sites including animal shelters and manholes within the study time and area. Informed by these preliminary findings, surveys were systematized in three fortnightly rounds during the study period. In the city, households with (n = 15) and without (n = 15) animal shelters were included (Extended Data Table 7). At DDU, two dormitory buildings which reported the highest number of malaria cases and their surroundings were selected. Adult mosquitoes were surveyed indoor, outdoor, in animal shelters, in overhead tanks and in manholes using Prokopack aspirators for 10 min between 6:00 and 8:00. Animal shelters were not available at DDU. Adult-caught mosquitoes (sorted on the basis of their abdominal status), and those raised from aquatic stages, were morphologically identified to the species level22 (Extended Data Table 8). Anopheles mosquitoes were individually preserved in tubes that contained silica gel desiccant in zipped bags and transported to the lab at the AHRI for further analysis. The global positioning system (GPS) coordinates of the households and immature and adult mosquito collection sites were recorded using GARMIN handheld GPS navigator (GARMIN GPSMAP 64S).
Laboratory proceduresNucleic acid extraction from whole blood and parasite quantification, and genotyping
Blood samples in ethylenediaminetetraacetic acid (EDTA) tubes were used to extract genomic DNA using MagMAX magnetic bead-based technology DNA multi-sample kit on KingFisher Flex robotic extractor machine (Thermo Fisher Scientific). Fifty microliters of whole blood input was eluted in a 150 μl low-salt elution buffer. Multiplex qPCR targeting the 18S rRNA small subunit gene for P. falciparum and P. vivax was run using primer and probe sequences described by Hermsen57 and Wampfler58 using TaqMan Fast Advanced Master Mix (Applied Biosystems). P. falciparum parasites were quantified using standard curves generated from a serial dilution of NF54 ring stage parasites (106 to 103 parasites ml−1). For P. vivax, parasite quantification was done using plasmid constructs to infer copy numbers by running serial dilutions (107 to 103 copies µl−1) of plasmids having the amplicon. Serial dilutions of the standard curves were generated in duplicate on each plate. Multiplexed amplicon sequencing was performed on qPCR-positive samples with reagents and protocol as in Tessema et al.59. DNA was amplified for 15 or 20 cycles in multiplexed PCR, depending on parasitemia and ability to amplify, and for 15 cycles for indexing PCR. The primer pools used in this study comprised high-diversity microhaplotype targets (n = 162), polymorphisms associated with drug resistance, and targets in and adjacent to pfhrp2 and pfhrp3 to assess for gene deletion (Primer pools 1A and 5 as described in protocols.io repository)60. Amplified libraries were sequenced in a NextSeq 2000 or a MiniSeq instrument using 150PE reads with 10% PhiX.
Nucleic acid extraction from mosquitoes, assessment of infectivity and bloodmeal source and confirmation of morphological species identification
Infection detection in wild caught mosquitoes is commonly based on an enzyme-linked immunosorbent assay (ELISA)-based protocol that targets circumsporzoite protein (CSP) that is expressed on the surface of Plasmodium sporozoites. Low-level expression of CSP at stages of sporogony before the parasites migrate to the salivary gland might interfere with signal detected61. Several studies have reported false positive results when targeting CSP especially in zoophilic mosquitoes62,63. The false positive results could lead to an overestimation of mosquito infection rates. To achieve a conservative estimate of mosquito infection rates, we implemented stringent steps as indicated below:
(1)
Bisected mosquitoes: We observed previously61 that a signal detected from an earlier stage of sporogony might interfere with interpretation of sporozoite detection, probably causing false positive results. We bisected the mosquitoes anterior to the thorax–abdomen junction under a stereo microscope before processing them for infection detection64. The head and thoraces were processed and stored separately from the abdomen of the mosquitoes. We only used the head and thorax part for infection detection following homogenization in a robust semi-high-throughput mini-bead beater protocol we developed previously65. The heads and thoraces of the mosquitoes were homogenized in 150 µl molecular-grade water that contains 0.2 g zirconium bead (1 mm diameter) using a Mini-Bead Beater 96. Part of the homogenate (50 µl) was used for nucleic acid extraction using cetyl trimethyl ammonium bromide62; 100 µl grinding buffer (0.5% w/v case in, 0.1 N NaOH in 10 mM PBS, pH 7.4, and 0.5% IGPAL CA-630) was added to the remaining that was used to screen samples for circumsporozoite in bead-based assay.
(2)
Circumsporozoite bead-based assay: We adopted the most advanced (highly sensitive) bead-based assay for infection detection in mosquitoes66 by targeting CSP. Antibody-coupled magnetic beads and biotinylated secondary antibodies were obtained from the Centers for Disease Control and Prevention, Division of Parasitic Diseases and Malaria, Entomology Branch, Atlanta, GA, USA, and implemented as described before7 and were run using MagPix immunoanalyzer (Luminex Corp, CN-0269-01).
(3)
Quality control to reduce cross-reactivity: The bead-based assay we adopted may eliminate false negatives due to lower limit of detection than previous ELISA-based assays66 but also brings a challenge of enhanced detection of cross-reacting proteins. To reduce this chance, mosquito homogenate was boiled at 100 °C before processing to eliminate false positives that may be caused by heat-unstable cross-reactive proteins to strengthen the validity of the results. To ascertain this specificity issue, we have included colony-maintained An. arabiensis and An. stephensi mosquitoes fed on sugar solution and patients’ blood in direct membrane feeding assays (had infection status determined morphologically in the same mosquito batches) that were used as negative and positive controls, respectively. Plasmodium-infected mosquitoes were used as positive controls along with sugar-fed mosquitoes as negative controls in every extraction round (Supplementary Fig. 3 and Supplementary Tables 1 and 2).
(4)
Retesting and confirmatory 18S-based species-specific PCR: Samples with higher mean fluorescence intensity signal than the negative controls plus three standard deviations and a representative set of mosquitoes that gave low signal were rerun to confirm the observations. Genomic DNA extracted from the head and thoraces of all mosquitoes was tested on a PCR that targeted 18S small ribosomal subunit gene as a confirmatory test. Only mosquito samples positive by the CSP-based assays and 18S-based PCR were considered infected.
Nucleic acid was extracted from the abdomen of fully engorged mosquitoes for bloodmeal source identification following the same procedure used for the head and thoraces using a cetyltrimethylammonium bromide (CTAB)-based method as described before67. A multiplex PCR assay that amplifies the cytochrome b gene based on Kent and Norris68 was used for bloodmeal source analysis. We have introduced slight modifications to improve product size separation on gel electrophoresis. The multiplex of cow and dog was separately done from the multiplex of goat and human. The optimized PCR thermal cycler conditions were: 5 min at 95 °C as an initial denaturation followed by 40 cycles of denaturation at 95 °C for 60 s, annealing at 56 °C for 60 s for cow and dog multiplex, and 62 °C for goat and human multiplex, followed by an extension at 72 °C for 60 s, and 1 cycle of the final extension at 72 °C for 7 min.
Confirmation of the Anopheles morphological identification was done following a recently published protocol that targets the ITS2 gene69. An. stephensi diagnostic amplicon of 438 bp size was expected while a universal amplicon of varying sizes (>600 bp), depending on the length of ITS2 in a particular species, was expected in this multiplex protocol (Supplementary Fig. 4). The universal amplicon was used to serve as an internal control to rule out PCR failure.
Data management and analysisData management
Study data collection tools (mobile application version 5.20.11) were prepared and managed using REDCap electronic data capture tools hosted at AHRI. CSV files exported from REDCap were analyzed using STATA 17 (StataCorp), RStudio v.2022.12.0.353 (Posit, 2023), QGIS v.3.22.16 (QGIS Development Team, 2023, QGIS Geographic Information System, Open Source Geospatial Foundation Project), GraphPad Prism 5.03 (GraphPad Software) and RStudio using packages lme4 (generalized linear mixed models) and dcifer70 (pairwise relatedness analysis on P. falciparum genotypes in diverse loci).
Description of study variables
We collected the following variables in this study:
Sociodemographic: sex, age, educational level and occupation
Household characteristics: main materials used for building the household, fuel source, water source and presence of water bodies near the household/dormitory, and presence of livestock
Intervention: presence, number, and condition of bed nets, use of bed nets, use of smoke repellents or aerosol mosquito spray, and history of insecticide residual spray
Diagnosis and treatment: malaria test result by RDTs and microscopy, temperature, presence of symptoms and treatment history, and pregnancy status
Human behavior: travel history, health seeking behavior, sleeping and waking time, and sleeping place
Entomological survey: mosquito collection method and time of collection, mosquito species detected and density, Anopheles species detected and density, abdominal status of mosquitoes detected, type of aquatic habitat near the household/dormitory, and type and characteristics of water sources detected within 100-m radius around the household/dormitory
Bioinformatic analysis
FASTQ files from multiplexed amplicon sequencing of P. falciparum were subjected to filtering, demultiplexing and allele inference using a Nextflow-based pipeline71. We used cut adapt to demultiplex reads for each locus based on the locus primer sequences (no mismatches or indels allowed), filter reads by length (100 base pairs) and quality (default NextSeq quality trimming). We used dada2 to infer variants and remove chimeras. Reads with a Phred quality score of less than 5 were truncated. The leftmost base was trimmed and trimmed reads of less than 75 base pairs were filtered out. Default values were used for all other parameters. We then aligned alleles to their reference sequence and filtered out reads with low alignment. We masked homopolymers and tandem repeats to avoid false positives.
Genetic analysis
Pairwise relatedness analysis was performed on P. falciparum genotypes in diverse loci using Dcifer with default settings70. Pairwise relatedness was only considered between samples where the lower 95% CI of estimated relatedness was greater than 0.1. Point estimates of pairwise relatedness that satisfied this threshold were then binned into low, medium and high relatedness at greater than 0.2, 0.5 and 0.9 respectively. Samples were then clustered based on pairwise relatedness. Drug resistance marker genotypes were extracted from loci of interest. Evidence of pfhrp2 and pfhrp3 deletions were identified from a drop in normalized coverage in amplicons within and surrounding pfhrp2 and pfhrp3. Complexity of infection was estimated by taking the 0.97 quantile (fifth highest number) of observed alleles across loci to minimize the impact of false positives on estimates.
Epidemiological analysis
We used standard case–control analyses to examine the association between risk factors and malaria infection. It calculates point estimates and CIs for the OR along with the significance level based on the chi-squared test. Continuous variables were presented as median and IQR. Tests of association between two categorical variables were performed using chi-squared test on contingency tables. Mann–Kendall statistical test was used to test for monotonic (increasing or decreasing) trends of malaria cases using the secondary data obtained from the private and public health facilities at the city and DDU.
Spatial data analysis
As the dormitories within the university study site were located within a small area (20 buildings in 45,450 m2 area), clustering of prevalence data was assessed in the city only. The prevalence of malaria by RDT and/or microscopy was calculated for each household. Global and local Moran’s I calculations were used to estimate the level of spatial autocorrelation within household prevalence data. The statistical strength of association for global Moran’s I was calculated using Monte-Carlo methods based on 9,999 times permutations of the prevalence data. The Euclidean distance from the river to every site where adult or larval An. stephensi were located were calculated in meters.
Statistical analysis
To identify the association of An. stephensi and other risk factors for malaria positivity and quantify the variation in a parasite positive outcome in Dire Dawa, we employed a multilevel logistic regression model with nested random effects (heterogeneous household and case–control group variances) to account for intra-class correlation72. The covariates included for the multi-level logistic regression analysis with random effect are listed in detail in Supplementary Table 3. Having more than 30 potential covariates associated to malaria, more than one billion models for exhaustive best model searching (excluding interactions between covariates), we reduced the number of covariates to a manageable size by considering univariate generalized mixed models (with case index as random effect instead of setting which were not contributing to the differences in malaria positivity for cases and controls) and considering only the covariates with P value lower than 0.3 within these models (Supplementary Table 3). The decision to use case/control as random effect instead of fixed effect came from preliminary analysis that considered the best candidate(s) for random effects. Variable selection was performed by testing 2,000+ binomial logistic mixed models (number of tested models depending on initial screening). During the initial screening, a candidate variable was selected if its P value, obtained from a Wald test applied to the variable’s estimated coefficient in logistic regression, was lower than 0.3. The models were ranked on the basis of their Akaike Information Criteria (AIC) and the Bayes information criteria (BIC) values, with the top model being the one with the lowest AIC value73. Variable selection was repeated for three different response variables: model 1 with response RDT/microscopy, model 2 with response RDT/microscopy/qPCR, and, finally, model 3 with response qPCR. As a result, only five of the 12 factors assessed for individual and household characteristics (sex, age, An. stephensi larvae and/or adult presence, natural waterbody existence, and use of aerosol insecticide spray) were included for the final model (Supplementary Table 4). We also explored interactions between gender, age and site.
After model selection with several model outcomes and distribution (Supplementary Table 4), the binomial model with outcome represented by malaria positivity (positive/negative) using RDT and/or microscopy best represented the relationship between malaria and risk factors (Supplementary Table 4)74. In this model, the employment of geographic unit’s effects such as household and area setting (city versus university) enabled us to control for unknown variations by including them as random effects in the model. In fact, individuals living in the same household may share exposures that can determine similarities in malaria transmission as well as in the larger setting (city versus university).
Let yij denote the malaria outcome of the ith individual in the jth household or cluster, identified by the RDT and/or microscopy with probability πij where yij = 1 denotes the individual tested positive, while yij = 0 denotes the individual tested negative for malaria. A multilevel logistic regression model with random effects for the outcome yij is given by
$$log it({pi }_{ij})={beta }_{0j}+beta {X}_{ij}+{u}_{j}$$
where Xij = (1, x1ij,…, xpij) is vector of p explanatory variables or covariates measured on the i individual and on the j household (cluster), β is vector of fixed regression coefficients or parameters and uj is a random effect varying over household and case control.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Source link : https://www.nature.com/articles/s41591-023-02641-9
Author :
Publish date : 2023-10-26 07:00:00
Copyright for syndicated content belongs to the linked Source.