The present reveals the power of genomic surveillance for SARS-COV-2 in understanding the the dynamics in the epidemiology of COVID-19 at national level13,16. In settings with limited access to sequencing17,18, collaborative efforts enabled sampling and sequencing of SARS-CoV-2 through the genomic surveillance platform in place19,20,21, with international partnerships13,16,22. Thus, this initiative could be viable for any genomic surveillance of any pathogen of pandemic or epidemic potential (Ebola, Zika, Mpox viruses, cholera, antimicrobial resistant strains, etc.) as supported by Africa CDC3 and other agencies18,23.
In this study, the mean age of the population was 36 (26–45) years. This represents the most active population often involved in travels, social or occupational activities that increase exposure to SARS-CoV-2, as previously reported in similar settings24,25,26. However, this observation is different in the Western world most likely due to greater adherence to barrier measures27. The sex ratio showed a similar distribution in SARS-CoV-2 cases, indicating similar risk of infection/exposure at population-level28.
Sampling was from 9/10 of the national regions, suggesting wide near-national coverage (only North-West region excluded). However, only 30% of regions achieved a desirable sampling for genomic surveillance (at least 10% of sequence data). This is in line with coverage of molecular testing mostly found in major townships25,29. Thus, genomic data from difficult-to-reach settings are less covered30.
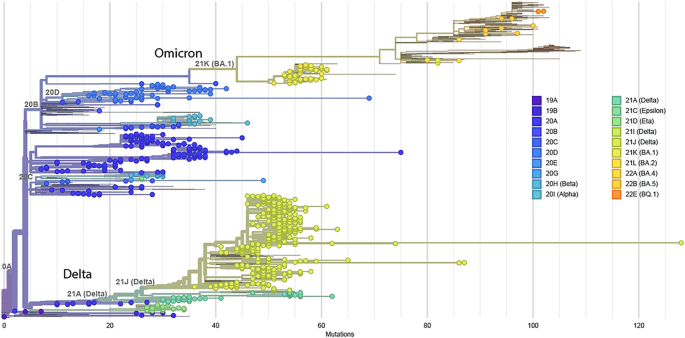
According to available sequence data, the viral sub-lineages of origin and non-VOCs represent the majority (74%) as compared to VOCs, reflecting efforts in genomic surveillance during the early phase of the pandemic even at the global level31,32,33. Interestingly, transmission dynamics confirms the first cases as viral sub-lineages of origin; followed between December 2020 and April 2021 by the co-introduction of alpha and beta as first VOCs, favoured by population migrations for end-of-year holidays from the Western world where these variants were already prevalent31,34. First cases of delta were identified in March 2021 with a peak in October 2021. The increase in cases with delta would be favoured by the variant affinity for the ACE2 receptor due to mutations35, leading to enhanced viral attachment, high viral load and prolonged duration of infection36. First cases of omicron were found in September 2021 and predominate as from December 2021 to reach 100% by end of February 2022. Regarding the evolutionary trends of variants per wave, the first wave (driven by lineages of origin) has a medium length in duration, a moderate number of cases and deaths, but a high CFR which could be attributed to limited experience/logistics of the health system to respond to an unknown disease37; not neglecting the effect of stigma and fear on late clinic attendance during that early phase38,39. The second wave was driven by the co-introduction of alpha and beta variants alongside other sub-lineages and non-VOCs, and was characterised by the longest outbreak duration of about five months, a high number of cases and hospitalisations, but a moderate CRF. The decreased CFR might be due to timely clinic attendance and gradual experience in case management39,40. Moreover, as compared to the high circulation of sub-lineages of origin and non-VOCs, alpha and beta variants had limited effects on transmission and disease severity, which prone their disappearance41,42. In contrast, the third wave (driven by the Delta variant) had a moderate duration of about three months but an increased/high number of confirmed cases (over 20,000), hospitalisations (over 1000) and high CFR (2%). Thus, in a relatively short timeframe, delta variant considerably increased both the transmission rate and the disease severity41,42. The fourth wave (driven solely by omicron) had a very short duration of about two months and a reduced/moderate number of cases (10,803) and low number of hospitalisations/deaths and CFR (0.73%), which underscores the contribution of omicron on viral transmission but without severity43,44. The overall decline in outbreak duration and number of cases across waves highlights the fact that the extent of viral transmission/spread was mainly driven by the duration of the outbreak. However, the low number of cases and high CFRs during wave 1 and wave 3 signifies that the original viral lineage and delta variant contributed to COVID-19 severity in the Cameroonian context, alongside other multifaceted determinants (naïve populations at the early stage of the pandemic, etc.)25. Thus, among VOCs, delta was a substantial driver of COVID-19 severity and death, likely attributed to loss in antibody affinity due to viral antigenic mutations in the receptor-binding domain45.
The main limitation of our study is the lack of proportionally representative samples across all regions and across the four waves. This suggests a reduced generalizability of the findings, especially in terms of overall prevailing viral lineages. Nonetheless, these findings, generated with high-quality and full-length sequences validated by a public repository (GISAID) and interpreted using a robust phylogenetic pipeline, provide evidence with major public health implications to prepare for future pandemics15.
In a nutshell, our genomic surveillance with full-length sequences reveals four VOCs (alpha, beta, delta, and omicron) in Cameroon across four different epidemiological waves. SARS-CoV-2 infection in Cameroon has been driven by the viral lineages of origin in wave 1, the co-introduction of alpha and beta variants in wave 2, delta variant in wave 3 and omicron variant in wave 4, with an overall declining trend in the wave duration, confirmed cases, hospitalisations and CFR over time. While viral transmission was not dependent on viral clades, SARS-CoV-2 viral sub-lineage of origin (at the early phase) and Delta variant appeared to be the drivers of COVID-19 severity in Cameroon.
Source link : https://www.nature.com/articles/s41598-023-48773-3
Author :
Publish date : 2023-12-08 08:00:00
Copyright for syndicated content belongs to the linked Source.